J Breast Cancer.
2018 Sep;21(3):297-305. 10.4048/jbc.2018.21.e38.
Expression of DNA Damage Response Proteins and Associations with Clinicopathologic Characteristics in Chinese Familial Breast Cancer Patients with BRCA1/2 Mutations
- Affiliations
-
- 1Department of Breast Surgery, Fudan University Shanghai Cancer Center, Shanghai, China. zhenhu@fudan.edu.cn
- 2Department of Pathology, Fudan University Shanghai Cancer Center, Shanghai, China.
- 3Department of Tissue Bank, Fudan University Shanghai Cancer Center, Shanghai, China.
- 4Department of Pathology, Shanghai Medical College, Fudan University, Shanghai, China.
- KMID: 2421370
- DOI: http://doi.org/10.4048/jbc.2018.21.e38
Abstract
- PURPOSE
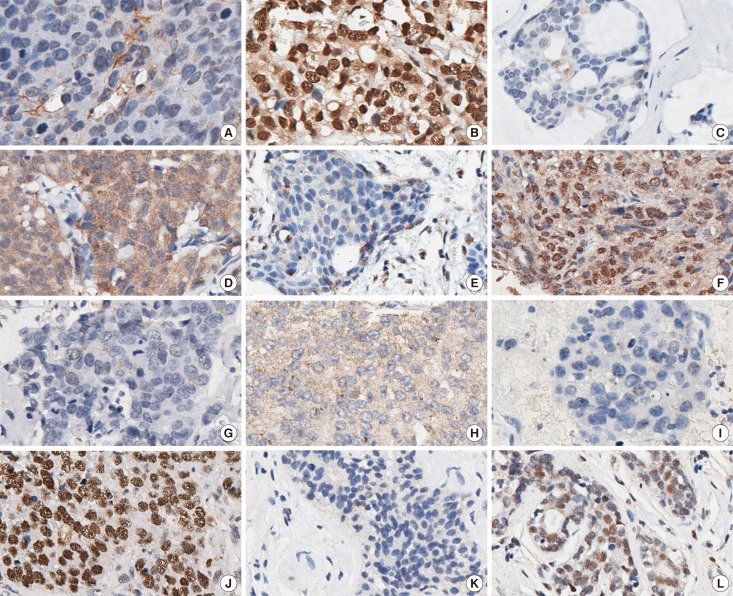
The characteristic expression of DNA damage response proteins in familial breast cancers with BRCA1, BRCA2, or non-BRCA1/2 mutations has not been analyzed in Chinese patients. Our study aimed to assess the differential expression of microcephalin 1 (BRIT1), ATM serine/threonine kinase (ATM), checkpoint kinase 2 (CHEK2), BRCA1, RAD51 recombinase (RAD51), and poly (ADP-ribose) polymerase 1 (PARP-1) and establish the profile of Chinese familial breast cancers with different mutation status.
METHODS
We constructed five tissue microarrays from 183 familial breast cancer patients (31 with BRCA1 mutations; 14 with BRCA2 mutations, and 138 with non-BRCA1/2 mutations). The DNA response and repair markers used for immunohistochemistry analysis included BRIT1, ATM, CHEK2, BRCA1, RAD51, and PARP-1. The expressions of these proteins were analyzed in BRCA1/2 mutated tumors. The association between pathologic characteristics with BRCA1/2 mutation status was also analyzed.
RESULTS
In familial breast cancer patients, BRCA1 mutated tumors were more frequent with high nuclear grade, estrogen receptor/progesterone receptor/human epidermal growth factor receptor 2 negative, low Ki-67, and positive CK5/6. BRCA1 mutated tumors had lower CHEK2 and higher cytoplasmic BRIT1 expression than BRCA2 and non-BRCA1/2 mutation tumors. BRCA2-associated tumors showed higher CHEK2 and cytoplasmic RAD51 expression than those in other groups. Nuclear PARP-1 expression in BRCA1/2-associated tumors was significantly higher than in non-BRCA1/2 mutation tumors. Moreover, we found quite a few of negative PARP-1 expression cases in BRCA1/2 mutated groups.
CONCLUSION
The clinicopathologic findings of BRCA1-associated Chinese familial breast cancers were similar to the results of other studies. Chinese familial breast cancer patients with BRCA1/2 mutations might have distinctive expression of different DNA damage response proteins. The reduced expression of PARP-1 in Chinese BRCA1/2 mutated breast cancer patients could influence the therapeutic outcome of PARP-1 inhibitors.
MeSH Terms
-
Asian Continental Ancestry Group*
Breast Neoplasms*
Breast*
Checkpoint Kinase 2
Cytoplasm
DNA Damage*
DNA Repair
DNA*
Estrogens
Genes, BRCA1
Genes, BRCA2
Humans
Immunohistochemistry
Phosphotransferases
Rad51 Recombinase
Receptor, Epidermal Growth Factor
DNA
Estrogens
Phosphotransferases
Rad51 Recombinase
Receptor, Epidermal Growth Factor
Figure
Reference
-
1. King MC, Marks JH, Mandell JB. New York Breast Cancer Study Group. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science. 2003; 302:643–646. PMID: 14576434.2. Shih HA, Couch FJ, Nathanson KL, Blackwood MA, Rebbeck TR, Armstrong KA, et al. BRCA1 and BRCA2 mutation frequency in women evaluated in a breast cancer risk evaluation clinic. J Clin Oncol. 2002; 20:994–999. PMID: 11844822.
Article3. Yang X, Wu J, Lu J, Liu G, Di G, Chen C, et al. Identification of a comprehensive spectrum of genetic factors for hereditary breast cancer in a Chinese population by next-generation sequencing. PLoS One. 2015; 10:e0125571. PMID: 25927356.
Article4. Aleskandarany M, Caracappa D, Nolan CC, Macmillan RD, Ellis IO, Rakha EA, et al. DNA damage response markers are differentially expressed in BRCA-mutated breast cancers. Breast Cancer Res Treat. 2015; 150:81–90. PMID: 25690937.
Article5. Partipilo G, Simone G, Scattone A, Scarpi E, Azzariti A, Mangia A. Expression of proteins involved in DNA damage response in familial and sporadic breast cancer patients. Int J Cancer. 2016; 138:110–120. PMID: 26205471.
Article6. Richardson J, Shaaban AM, Kamal M, Alisary R, Walker C, Ellis IO, et al. Microcephalin is a new novel prognostic indicator in breast cancer associated with BRCA1 inactivation. Breast Cancer Res Treat. 2011; 127:639–648. PMID: 20632086.
Article7. Bartek J, Lukas J, Bartkova J. DNA damage response as an anti-cancer barrier: damage threshold and the concept of ‘conditional haploinsufficiency’. Cell Cycle. 2007; 6:2344–2347. PMID: 17700066.
Article8. Tommiska J, Bartkova J, Heinonen M, Hautala L, Kilpivaara O, Eerola H, et al. The DNA damage signalling kinase ATM is aberrantly reduced or lost in BRCA1/BRCA2-deficient and ER/PR/ERBB2-triple-negative breast cancer. Oncogene. 2008; 27:2501–2506. PMID: 17982490.
Article9. Lee JS, Collins KM, Brown AL, Lee CH, Chung JH. hCds1-mediated phosphorylation of BRCA1 regulates the DNA damage response. Nature. 2000; 404:201–204. PMID: 10724175.
Article10. Vahteristo P, Bartkova J, Eerola H, Syrjäkoski K, Ojala S, Kilpivaara O, et al. A CHEK2 genetic variant contributing to a substantial fraction of familial breast cancer. Am J Hum Genet. 2002; 71:432–438. PMID: 12094328.
Article11. Venkitaraman AR. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell. 2002; 108:171–182. PMID: 11832208.
Article12. Forget AL, Kowalczykowski SC. Single-molecule imaging brings Rad51 nucleoprotein filaments into focus. Trends Cell Biol. 2010; 20:269–276. PMID: 20299221.
Article13. Comen EA, Robson M. Inhibition of poly(ADP)-ribose polymerase as a therapeutic strategy for breast cancer. Oncology (Williston Park). 2010; 24:55–62. PMID: 20187322.14. Detre S, Saclani Jotti G, Dowsett M. A “quickscore” method for immunohistochemical semiquantitation: validation for oestrogen receptor in breast carcinomas. J Clin Pathol. 1995; 48:876–878. PMID: 7490328.
Article15. Domagala P, Huzarski T, Lubinski J, Gugala K, Domagala W. PARP-1 expression in breast cancer including BRCA1-associated, triple negative and basal-like tumors: possible implications for PARP-1 inhibitor therapy. Breast Cancer Res Treat. 2011; 127:861–869. PMID: 21409392.
Article16. Mego M, Cierna Z, Svetlovska D, Macak D, Machalekova K, Miskovska V, et al. PARP expression in germ cell tumours. J Clin Pathol. 2013; 66:607–612. PMID: 23486608.
Article17. Eerola H, Heikkilä P, Tamminen A, Aittomäki K, Blomqvist C, Nevanlinna H. Histopathological features of breast tumours in BRCA1, BRCA2 and mutation-negative breast cancer families. Breast Cancer Res. 2005; 7:R93–R100. PMID: 15642173.
Article18. Lakhani SR, Van De Vijver MJ, Jacquemier J, Anderson TJ, Osin PP, McGuffog L, et al. The pathology of familial breast cancer: predictive value of immunohistochemical markers estrogen receptor, progesterone receptor, HER-2, and p53 in patients with mutations in BRCA1 and BRCA2. J Clin Oncol. 2002; 20:2310–2318. PMID: 11981002.
Article19. Lakhani SR, Reis-Filho JS, Fulford L, Penault-Llorca F, van der Vijver M, Parry S, et al. Prediction of BRCA1 status in patients with breast cancer using estrogen receptor and basal phenotype. Clin Cancer Res. 2005; 11:5175–5180. PMID: 16033833.
Article20. Mangia A, Chiriatti A, Tommasi S, Menolascina F, Petroni S, Zito FA, et al. BRCA1 expression and molecular alterations in familial breast cancer. Histol Histopathol. 2009; 24:69–76. PMID: 19012246.21. Yoshikawa K, Honda K, Inamoto T, Shinohara H, Yamauchi A, Suga K, et al. Reduction of BRCA1 protein expression in Japanese sporadic breast carcinomas and its frequent loss in BRCA1-associated cases. Clin Cancer Res. 1999; 5:1249–1261. PMID: 10389907.22. Magdinier F, Ribieras S, Lenoir GM, Frappart L, Dante R. Down-regulation of BRCA1 in human sporadic breast cancer; analysis of DNA methylation patterns of the putative promoter region. Oncogene. 1998; 17:3169–3176. PMID: 9872332.
Article23. Bianco T, Chenevix-Trench G, Walsh DC, Cooper JE, Dobrovic A. Tumour-specific distribution of BRCA1 promoter region methylation supports a pathogenetic role in breast and ovarian cancer. Carcinogenesis. 2000; 21:147–151. PMID: 10657950.
Article24. Bahassi EM, Ovesen JL, Riesenberg AL, Bernstein WZ, Hasty PE, Stambrook PJ. The checkpoint kinases Chk1 and Chk2 regulate the functional associations between hBRCA2 and Rad51 in response to DNA damage. Oncogene. 2008; 27:3977–3985. PMID: 18317453.
Article25. Abdel-Fatah TM, Arora A, Alsubhi N, Agarwal D, Moseley PM, Perry C, et al. Clinicopathological significance of ATM-Chk2 expression in sporadic breast cancers: a comprehensive analysis in large cohorts. Neoplasia. 2014; 16:982–991. PMID: 25425972.
Article26. Davies AA, Masson JY, McIlwraith MJ, Stasiak AZ, Stasiak A, Venkitaraman AR, et al. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol Cell. 2001; 7:273–282. PMID: 11239456.
Article27. Honrado E, Osorio A, Palacios J, Milne RL, Sánchez L, Díez O, et al. Immunohistochemical expression of DNA repair proteins in familial breast cancer differentiate BRCA2-associated tumors. J Clin Oncol. 2005; 23:7503–7511. PMID: 16234517.
Article28. Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006; 7:517–528. PMID: 16829982.
Article29. Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009; 361:123–134. PMID: 19553641.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- DNA Damage Response Mediated through BRCA1
- Novel Germline Mutations of BRCA1 and BRCA2 in Korean Familial Breast Cancer Patients
- Frequency of BRCA1 and BRCA2 Germline Mutations Detected by Protein Truncation Test and Cumulative Risks of Breast and Ovarian Cancer among Mutation Carriers in Japanese Breast Cancer Families
- The Expression of ERCC1, RRM1, and BRCA1 in Breast Cancer According to the Immunohistochemical Phenotypes
- A Study for Germline Mutation of BRCA1 in Early Onset Breast Cancer Patients