J Korean Med Sci.
2011 Mar;26(3):352-359. 10.3346/jkms.2011.26.3.352.
The Expression of ERCC1, RRM1, and BRCA1 in Breast Cancer According to the Immunohistochemical Phenotypes
- Affiliations
-
- 1Department of Pathology, Yonsei University College of Medicine, Seoul, Korea. kjs1976@yuhs.ac
- KMID: 2157871
- DOI: http://doi.org/10.3346/jkms.2011.26.3.352
Abstract
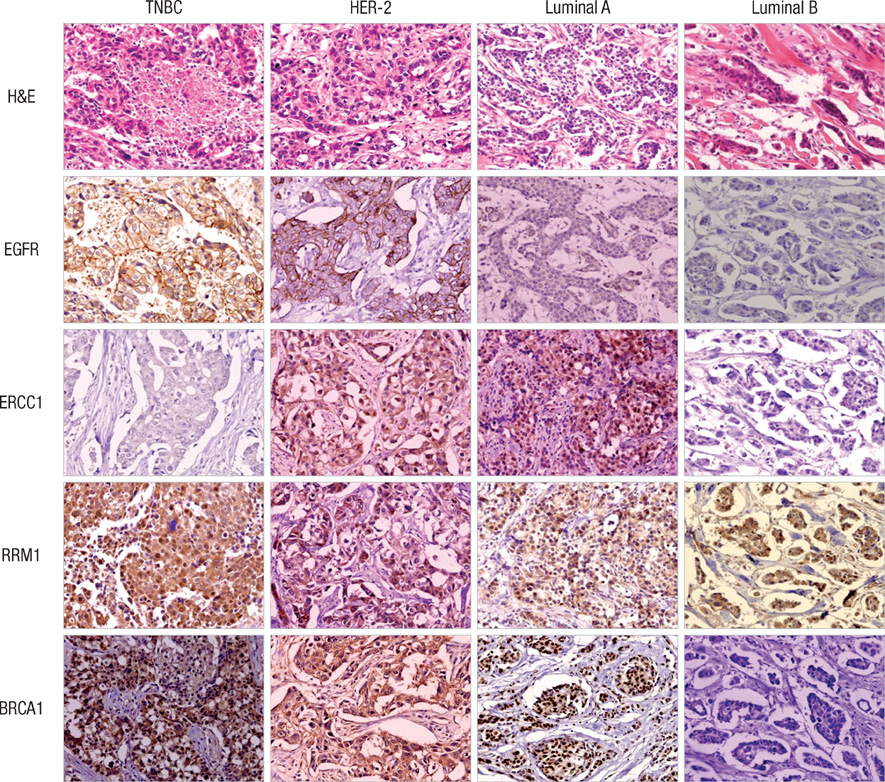
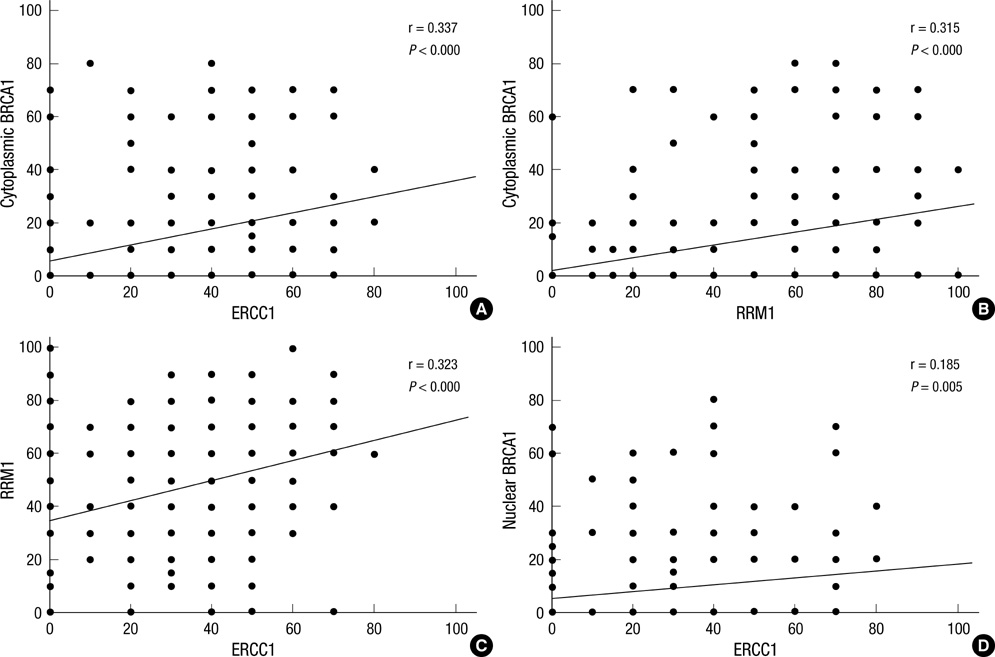
- We studied the expression of BRCA1, ERCC1, and RRM1 which play an important role in DNA repair systems in breast cancer. Immunohistochemical staining for EGFR, BRCA1, ERCC1, and RRM1 were performed by using a tissue microarray made from 230 breast cancer patients. Patients were classified into luminal A, luminal B, HER-2, and triple negative breast cancer (TNBC) types according to ER, PR, and HER-2 expression. The expression of ERCC1, RRM1, and BRCA1 were correlated (P < 0.05). The expression level of ERCC1 was the lowest in TNBC type (P = 0.031), ERCC1 negativity was more prominent in TNBC and luminal B groups than luminal A and HER-2 groups (P = 0.013). Cases with EGFR overexpression showed high expression of RRM1 and BRCA1 (P = 0.046, and 0.004, respectively). In conclusion, the expression of ERCC1 is particularly lower in TNBCs than other types of breast cancers.
Keyword
MeSH Terms
-
Adult
BRCA1 Protein/*genetics/metabolism
Breast Neoplasms/*genetics/metabolism/pathology
DNA Repair
DNA-Binding Proteins/*genetics/metabolism
Disease-Free Survival
Endonucleases/*genetics/metabolism
Female
Gene Expression
Humans
Immunohistochemistry
Middle Aged
Phenotype
Prognosis
Protein Array Analysis
Receptor, Epidermal Growth Factor/genetics/metabolism
Tumor Markers, Biological/*genetics/metabolism
Tumor Suppressor Proteins/*genetics/metabolism
Figure
Reference
-
1. Bhattacharyya A, Ear US, Koller BH, Weichselbaum RR, Bishop DK. The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. J Biol Chem. 2000. 275:23899–23903.2. Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, Martin NM, Jackson SP, Smith GC, Ashworth A. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005. 434:917–921.3. Nielsen TO, Hsu FD, Jensen K, Cheang M, Karaca G, Hu Z, Hernandez-Boussard T, Livasy C, Cowan D, Dressler L, Akslen LA, Ragaz J, Gown AM, Gilks CB, van de Rijn M, Perou CM. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004. 10:5367–5374.4. Zdraveski ZZ, Mello JA, Marinus MG, Essigmann JM. Multiple pathways of recombination define cellular responses to cisplatin. Chem Biol. 2000. 7:39–50.5. Bergamaschi A, Kim YH, Wang P, Sørlie T, Hernandez-Boussard T, Lonning PE, Tibshirani R, Børresen-Dale AL, Pollack JR. Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes Chromosomes Cancer. 2006. 45:1033–1040.6. Kennedy RD, Quinn JE, Mullan PB, Johnston PG, Harkin DP. The role of BRCA1 in the cellular response to chemotherapy. J Natl Cancer Inst. 2004. 96:1659–1668.7. Couch FJ, DeShano ML, Blackwood MA, Calzone K, Stopfer J, Campeau L, Ganguly A, Rebbeck T, Weber BL. BRCA1 mutations in women attending clinics that evaluate the risk of breast cancer. N Engl J Med. 1997. 336:1409–1415.8. Futreal PA, Liu Q, Shattuck-Eidens D, Cochran C, Harshman K, Tavtigian S, Bennett LM, Haugen-Strano A, Swensen J, Miki Y, Eddington K, McClure M, Frye C, Weaver-Feldhaus J, Ding W, Gholami Z, Soderkvist P, Terry L, Jhanwar S, Berchuck A, Iglehart JD, Marks J, Ballinger DG, Barrett JC, Skolnick MH, Kamb A, Wiseman R. BRCA1 mutations in primary breast and ovarian carcinomas. Science. 1994. 266:120–122.9. Foulkes WD, Brunet JS, Stefansson IM, Straume O, Chappuis PO, Bégin LR, Hamel N, Goffin JR, Wong N, Trudel M, Kapusta L, Porter P, Akslen LA. The prognostic implication of the basal-like (cyclin E high/p27 low/p53+/glomeruloid-microvascular-proliferation+) phenotype of BRCA1-related breast cancer. Cancer Res. 2004. 64:830–835.10. Turner N, Tutt A, Ashworth A. Hallmarks of 'BRCAness' in sporadic cancers. Nat Rev Cancer. 2004. 4:814–819.11. Abd El-Rehim DM, Ball G, Pinder SE, Rakha E, Paish C, Robertson JF, Macmillan D, Blamey RW, Ellis IO. High-throughput protein expression analysis using tissue microarray technology of a large well-characterised series identifies biologically distinct classes of breast cancer confirming recent cDNA expression analyses. Int J Cancer. 2005. 116:340–350.12. Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991. 19:403–410.13. Cutler SJ, Black MM, Mork T, Harvei S, Freeman C. Further observations on prognostic factors in cancer of the female breast. Cancer. 1969. 24:653–667.14. Rakha EA, El-Sayed ME, Green AR, Lee AH, Robertson JF, Ellis IO. Prognostic markers in triple-negative breast cancer. Cancer. 2007. 109:25–32.15. Tischkowitz M, Brunet JS, Bégin LR, Huntsman DG, Cheang MC, Akslen LA, Nielsen TO, Foulkes WD. Use of immunohistochemical markers can refine prognosis in triple negative breast cancer. BMC Cancer. 2007. 7:134.16. Rouzier R, Perou CM, Symmans WF, Ibrahim N, Cristofanilli M, Anderson K, Hess KR, Stec J, Ayers M, Wagner P, Morandi P, Fan C, Rabiul I, Ross JS, Hortobagyi GN, Pusztai L. Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clin Cancer Res. 2005. 11:5678–5685.17. Sotiriou C, Neo SY, McShane LM, Korn EL, Long PM, Jazaeri A, Martiat P, Fox SB, Harris AL, Liu ET. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc Natl Acad Sci USA. 2003. 100:10393–10398.18. Reardon JT, Vaisman A, Chaney SG, Sancar A. Efficient nucleotide excision repair of cisplatin, oxaliplatin, and Bis-aceto-ammine-dichloro-cyclohexylamine-platinum(IV) (JM216) platinum intrastrand DNA diadducts. Cancer Res. 1999. 59:3968–3971.19. Metzger R, Leichman CG, Danenberg KD, Danenberg PV, Lenz HJ, Hayashi K, Groshen S, Salonga D, Cohen H, Laine L, Crookes P, Silberman H, Baranda J, Konda B, Leichman L. ERCC1 mRNA levels complement thymidylate synthase mRNA levels in predicting response and survival for gastric cancer patients receiving combination cisplatin and fluorouracil chemotherapy. J Clin Oncol. 1998. 16:309–316.20. Shirota Y, Stoehlmacher J, Brabender J, Xiong YP, Uetake H, Danenberg KD, Groshen S, Tsao-Wei DD, Danenberg PV, Lenz HJ. ERCC1 and thymidylate synthase mRNA levels predict survival for colorectal cancer patients receiving combination oxaliplatin and fluorouracil chemotherapy. J Clin Oncol. 2001. 19:4298–4304.21. Niedernhofer LJ, Odijk H, Budzowska M, van Drunen E, Maas A, Theil AF, de Wit J, Jaspers NG, Beverloo HB, Hoeijmakers JH, Kanaar R. The structure-specific endonuclease Ercc1-Xpf is required to resolve DNA interstrand cross-link-induced double-strand breaks. Mol Cell Biol. 2004. 24:5776–5787.22. Li Q, Yu JJ, Mu C, Yunmbam MK, Slavsky D, Cross CL, Bostick-Bruton F, Reed E. Association between the level of ERCC-1 expression and the repair of cisplatin-induced DNA damage in human ovarian cancer cells. Anticancer Res. 2000. 20:645–652.23. Ceppi P, Volante M, Novello S, Rapa I, Danenberg KD, Danenberg PV, Cambieri A, Selvaggi G, Saviozzi S, Calogero R, Papotti M, Scagliotti GV. ERCC1 and RRM1 gene expressions but not EGFR are predictive of shorter survival in advanced non-small-cell lung cancer treated with cisplatin and gemcitabine. Ann Oncol. 2006. 17:1818–1825.24. Metro G, Zheng Z, Fabi A, Schell M, Antoniani B, Mottolese M, Monteiro AN, Vici P, Lara Rivera S, Boulware D, Cognetti F, Bepler G. In situ protein expression of RRM1, ERCC1, and BRCA1 in metastatic breast cancer patients treated with gemcitabine-based chemotherapy. Cancer Invest. 2010. 28:172–180.25. Tassone P, Di Martino MT, Ventura M, Pietragalla A, Cucinotto I, Calimeri T, Bulotta A, Neri P, Caraglia M, Tagliaferri P. Loss of BRCA1 function increases the antitumor activity of cisplatin against human breast cancer xenografts in vivo. Cancer Biol Ther. 2009. 8:648–653.26. Silver DP, Richardson AL, Eklund AC, Wang ZC, Szallasi Z, Li Q, Juul N, Leong CO, Calogrias D, Buraimoh A, Fatima A, Gelman RS, Ryan PD, Tung NM, De Nicolo A, Ganesan S, Miron A, Colin C, Sgroi DC, Ellisen LW, Winer EP, Garber JE. Efficacy of neoadjuvant Cisplatin in triple-negative breast cancer. J Clin Oncol. 2010. 28:1145–1153.27. Al-Mulla F, Abdulrahman M, Varadharaj G, Akhter N, Anim JT. BRCA1 gene expression in breast cancer: a correlative study between real-time RT-PCR and immunohistochemistry. J Histochem Cytochem. 2005. 53:621–629.28. Pérez-Vallés A, Martorell-Cebollada M, Nogueira-Vázquez E, García-García JA, Fuster-Diana E. The usefulness of antibodies to the BRCA1 protein in detecting the mutated BRCA1 gene. An immunohistochemical study. J Clin Pathol. 2001. 54:476–480.29. Yacoub A, McKinstry R, Hinman D, Chung T, Dent P, Hagan MP. Epidermal growth factor and ionizing radiation up-regulate the DNA repair genes XRCC1 and ERCC1 in DU145 and LNCaP prostate carcinoma through MAPK signaling. Radiat Res. 2003. 159:439–452.30. Rakha EA, El-Sheikh SE, Kandil MA, El-Sayed ME, Green AR, Ellis IO. Expression of BRCA1 protein in breast cancer and its prognostic significance. Hum Pathol. 2008. 39:857–865.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- A Study on BRCA1/2 Mutations, Hormone Status and HER-2 Status in Korean Women with Early-onset Breast Cancer
- Frequency of BRCA1 and BRCA2 Germline Mutations Detected by Protein Truncation Test and Cumulative Risks of Breast and Ovarian Cancer among Mutation Carriers in Japanese Breast Cancer Families
- Expression of BRCA1 Transcripts and Protein in Sporadic Ovarian Cancer
- A Study for Germline Mutation of BRCA1 in Early Onset Breast Cancer Patients
- Distribution of BRCA1 and BRCA2 Mutations in Asian Patients with Breast Cancer