Infect Chemother.
2018 Mar;50(1):29-37. 10.3947/ic.2018.50.1.29.
Genotypic Diversity of Multidrug Resistant Shigella species from Iran
- Affiliations
-
- 1Immunology Research Center, Tabriz University of Medical Sciences, Tabriz, Iran. rezaee@tbzmed.ac.ir
- 2Department of Microbiology, Faculty of Medicine, Tabriz University of Medical Sciences, Tabriz, Iran.
- 3Student Research Committee, Tabriz University of Medical Sciences, Tabriz, Iran.
- 4Iranian Social Security Organization, Emam Reza Hospital, Urmia, Iran.
- 5Infectious and Tropical Diseases Research Center, Tabriz University of Medical Sciences, Tabriz, Iran.
- 6Department of Microbiology and Virology, School of Medicine, Kerman University of Medical Sciences, Kerman, Iran.
- KMID: 2407965
- DOI: http://doi.org/10.3947/ic.2018.50.1.29
Abstract
- BACKGROUND
In many developing countries, shigellosis is endemic and also occurs in epidemics and treatment of multidrug-resistant (MDR) isolates are important. The aims of this study were to determine the antimicrobial susceptibility, prevalence of class 1 and 2 integrons and the clonal relatedness of isolates.
MATERIALS AND METHODS
Antimicrobial susceptibility tests were performed by disc diffusion method. Polymerase chain reaction (PCR)-sequencing technique was employed for detection and characterization of integrons. The genetic relatedness was evaluated by using enterobacterial repetitive intergenic consensus (ERIC) PCR.
RESULTS
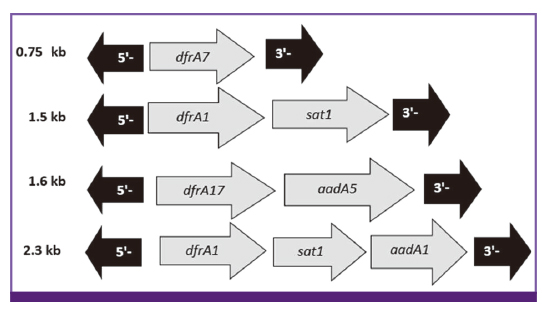
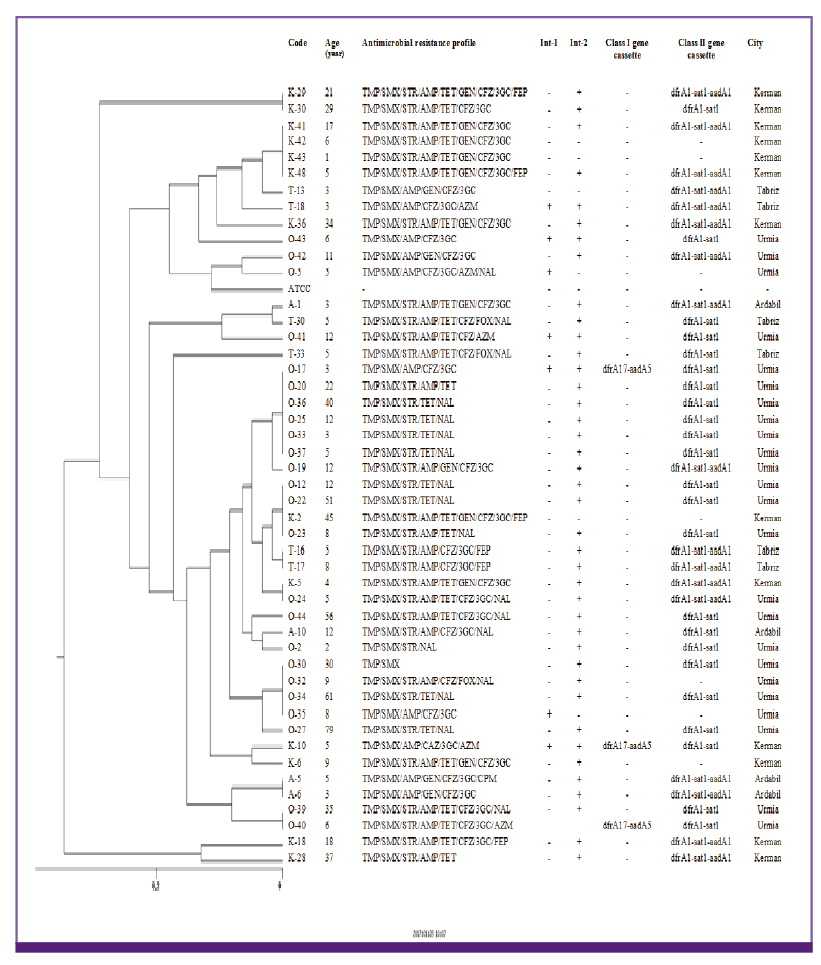
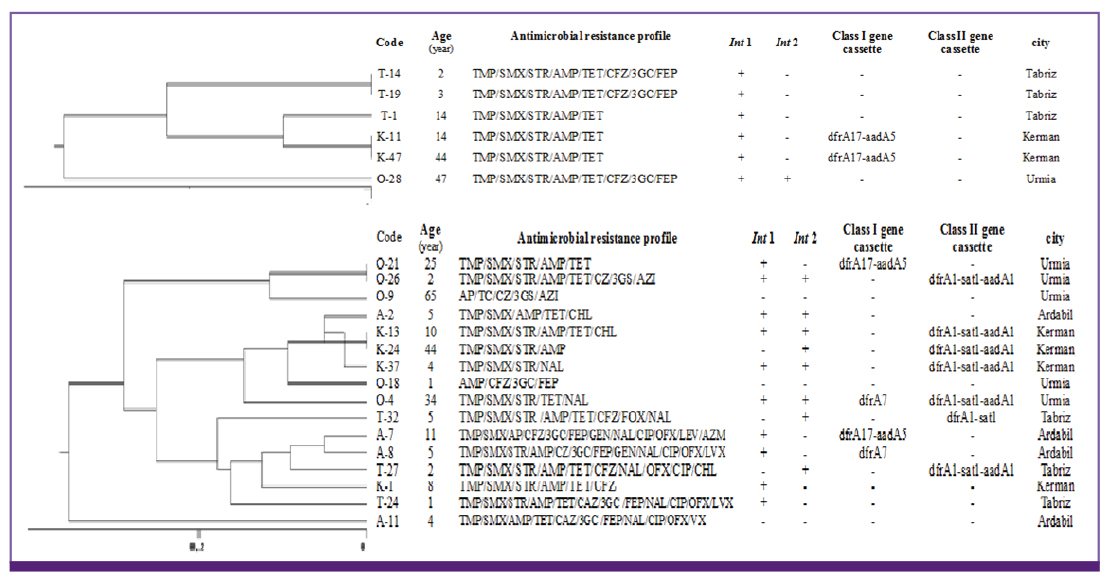
There was a high percentage of resistance to trimethoprim-sulfamethoxazole (TMP/SMX) (93.7%), ampicillin (AMP) (87.3%), streptomycin (STR) (84.5%) and tetracycline (TET) (78.9%). Multidrug resistant phenotype was seen in 95.1% of total isolates. Most common MDR profile was TMP/SMX/STR/AMP resistant pattern. Among the 142 Shigella spp. analyzed in this study, 28 isolates were positive for class 1 integron with two types of gene cassette arrays (dfrA17/aadA5 = 31.7% and dfrA7 = 3.8%). The class 2 integron was more frequently detected among the isolates (94.7%) with dfrA1/sat1/aadA1 (69.4%) and dfrA1/sat1 (30.6%) gene cassettes. ERIC-PCR results showed 6, 5, 4 and 3 main genotypes among S. flexneri, S. sonnei, S. boydii and S. dysenteriae isolates, respectively.
CONCLUSIONS
Our findings revealed that multidrug resistant Shigella species with high prevalence of class 2 integron were very common in Iran. In addition, ERIC-PCR patterns showed limited variety of clones are responsible for shigellosis in the region of the study.
Keyword
MeSH Terms
-
Ampicillin
Clone Cells
Consensus
Developing Countries
Diffusion
Dysentery, Bacillary
Genotype
Integrons
Iran*
Methods
Phenotype
Polymerase Chain Reaction
Prevalence
Shigella*
Streptomycin
Tetracycline
Trimethoprim, Sulfamethoxazole Drug Combination
Ampicillin
Streptomycin
Tetracycline
Trimethoprim, Sulfamethoxazole Drug Combination
Figure
Reference
-
1. Ahmed AM, Shimamoto T. Molecular characterization of multidrug-resistant Shigella spp. of food origin. Int J Food Microbiol. 2015; 194:78–82.
Article2. Tajbakhsh M, García Migura L, Rahbar M, Svendsen CA, Mohammadzadeh M, Zali MR, Aarestrup FM, Hendriksen RS. Antimicrobial-resistant Shigella infections from Iran: an overlooked problem? J Antimicrob Chemother. 2012; 67:1128–1133.
Article3. Pan JC, Ye R, Meng DM, Zhang W, Wang HQ, Liu KZ. Molecular characteristics of class 1 and class 2 integrons and their relationships to antibiotic resistance in clinical isolates of Shigella sonnei and Shigella flexneri . J Antimicrob Chemother. 2006; 58:288–296.
Article4. Ranjbar R, Mirsaeed Ghazi F. Antibiotic sensitivity patterns and molecular typing of Shigella sonnei strains using ERIC-PCR. Iran J Public Health. 2013; 42:1151–1157.5. Memish ZA, Venkatesh S, Shibl AM. Impact of travel on international spread of antimicrobial resistance. Int J Antimicrob Agents. 2003; 21:135–142.
Article6. Perilla MJ, Ajello G, Bopp C, Elliott J, Facklam R, Knapp JS, Popovic T, Wells J, Dowell SF. Manual for the laboratory identification and antimicrobial susceptibility testing of bacterial pathogens of public health importance in the developing world. Atlanta, GA: CDC;2003.7. Guardabassi L, Dijkshoorn L, Collard JM, Olsen JE, Dalsgaard A. Distribution and in-vitro transfer of tetracycline resistance determinants in clinical and aquatic Acinetobacter strains. J Med Microbiol. 2000; 49:929–936.
Article8. Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing. 25th informational supplement. Document M100-S25. Wayne, PA: CLSI;2015.9. Rezaee MA, Abdinia B, Abri R, Kafil HS. Comparison of the antibiotic resistance patterns among Shigella species isolated from pediatric hospital between 1995-1999 and 2009-2013 in North-West of Iran. J Anal Res Clin Med. 2014; 2:118–122.10. Ahmed AM, Shimamoto T, Shimamoto T. Molecular characterization of multidrug-resistant avian pathogenic Escherichia coli isolated from septicemic broilers. Int J Med Microbiol. 2013; 303:475–483.
Article11. Tamang MD, Oh JY, Seol SY, Kang HY, Lee JC, Lee YC, Cho DT, Kim J. Emergence of multidrug-resistant Salmonella enterica serovar Typhi associated with a class 1 integron carrying the dfrA7 gene cassette in Nepal. Int J Antimicrob Agents. 2007; 30:330–335.
Article12. Mazel D, Dychinco B, Webb VA, Davies J. Antibiotic resistance in the ECOR collection: integrons and identification of a novel aad gene. Antimicrob Agents Chemother. 2000; 44:1568–1574.
Article13. Dalla-Costa LM, Irino K, Rodrigues J, Rivera IN, Trabulsi LR. Characterisation of diarrhoeagenic Escherichia coli clones by ribotyping and ERIC-PCR. J Med Microbiol. 1998; 47:227–234.
Article14. Khakabimamaghani S, Najafi A, Ranjbar R, Raam M. GelClust: a software tool for gel electrophoresis images analysis and dendrogram generation. Comput Methods Programs Biomed. 2013; 111:512–518.
Article15. Zhang J, Jin H, Hu J, Yuan Z, Shi W, Yang X, Xu X, Meng J. Antimicrobial resistance of Shigella spp. from humans in Shanghai, China, 2004–2011. Diagn Microbiol Infect Dis. 2014; 78:282–286.
Article16. Eftekhari N, Bakhshi B, Pourshafie MR, Zarbakhsh B, Rahbar M, Hajia M, Ghazvini K. Genetic diversity of Shigella spp. and their integron content. Foodborne Pathog Dis. 2013; 10:237–242.17. Alizadeh-Hesar M, Bakhshi B, Najar-Peerayeh S. Clonal dissemination of a single Shigella sonnei strain among Iranian children during Fall 2012 in Tehran, IR Iran. Infect Genet Evol. 2015; 34:260–266.
Article18. Mardaneh J, Poor SA, Afrugh P. Prevalence of Shigella species and antimicrobial resistance patterns of isolated strains from infected pediatrics in Tehran. Int J Enteric Pathog. 2013; 1:e10705.19. Young HK, Amyes S. Plasmid trimethoprim resistance in Vibrio cholerae: migration of the type I dihydrofolate reductase gene out of the Enterobacteriaceae. J Antimicrob Chemother. 1986; 17:697–703.
Article20. Najibi S, Bakhshi B, Fallahzad S, Pourshafie MR, Katouli M, Sattari M, Alebouyeh M, Tajbakhsh M. Distribution of class 1 integrons among enteropathogenic Escherichia coli . Can J Microbiol. 2012; 58:637–643.
Article21. Cui X, Yang C, Wang J, Liang B, Yi S, Li H, Liu H, Li P, Wu Z, Xie J, Jia L, Hao R, Wang L, Hua Y, Qiu S, Song H. Antimicrobial resistance of Shigella flexneri serotype 1b isolates in China. PLoS One. 2015; 10:e0129009.22. Tariq A, Haque A, Ali A, Bashir S, Habeeb MA, Salman M, Sarwar Y. Molecular profiling of antimicrobial resistance and integron association of multidrug-resistant clinical isolates of Shigella species from Faisalabad, Pakistan. Can J Microbiol. 2012; 58:1047–1054.
Article23. McIver CJ, White PA, Jones LA, Karagiannis T, Harkness J, Marriott D, Rawlinson WD. Epidemic strains of Shigella sonnei biotype g carrying integrons. J Clin Microbiol. 2002; 40:1538–1540.
Article24. Jové T, Da Re S, Denis F, Mazel D, Ploy MC. Inverse correlation between promoter strength and excision activity in class 1 integrons. PLoS Genet. 2010; 6:e1000793.
Article25. Paulsen IT, Littlejohn TG, Rådström P, Sundström L, Sköld O, Swedberg G, Skurray RA. The 3'conserved segment of integrons contains a gene associated with multidrug resistance to antiseptics and disinfectants. Antimicrob Agents Chemother. 1993; 37:761–768.
Article26. Peirano G, Agersø Y, Aarestrup FM, dos Prazeres Rodrigues D. Occurrence of integrons and resistance genes among sulphonamide-resistant Shigella spp. from Brazil. J Antimicrob Chemother. 2005; 55:301–305.
Article27. Jin YH, Oh YH, Jung JH, Kim SJ, Kim JA, Han KY, Kim MY, Park SG, Lee YK. Antimicrobial resistance patterns and characterization of integrons of Shigella sonnei isolates in Seoul, 1999-2008. J Microbiol. 2010; 48:236–242.
Article28. Das A, Natarajan M, Mandal J. The emergence of quinolone resistant Shigella sonnei, Pondicherry, India. PLoS One. 2016; 11:e0160290.
Article29. Terajima J, Tamura K, Hirose K, Izumiya H, Miyahara M, Konuma H, Watanabe H. A multi-prefectural outbreak of Shigella sonnei infections associated with eating oysters in Japan. Microbiol Immunol. 2004; 48:49–52.
Article30. Navia MM, Capitano L, Ruiz J, Vargas M, Urassa H, Schellemberg D, Gascon J, Vila J. Typing and characterization of mechanisms of resistance of Shigella spp. isolated from feces of children under 5 years of age from Ifakara, Tanzania. J Clin Microbiol. 1999; 37:3113–3117.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Infection of Extended-Spectrum β-Lactamase Producing Shigella flexneri in Children Attending a Childcare Center in Korea
- Asymptomatic Urinary Tract Infection Caused by Shigella sonnei
- Multidrug-resistant Tuberculosis Spondylitis: A Case Report
- Multidrug-resistant Tuberculosis
- Low Levels of Extensively Drug-resistant Tuberculosis among Multidrug Resistant Tuberculosis Isolates and Their Relationship to Risk Factors: Surveillance in Tehran, Iran; 2006 to 2014