Lab Anim Res.
2017 Jun;33(2):157-164. 10.5625/lar.2017.33.2.157.
Comparative study of fatty liver induced by methionine and choline-deficiency in C57BL/6N mice originating from three different sources
- Affiliations
-
- 1College of Pharmacy, Pusan National University, Busan, Korea. youngjung@pusan.ac.kr
- 2Department of Clinical Laboratory Science, College of Nursing and Healthcare Science, Dong-Eui University, Busan, Korea.
- 3College of Pharmacy, Kyungsung University, Busan, Korea.
- 4Department of Biomedical Laboratory Science, Daekyeung College, Gyeongsan, Korea.
- 5Department of Microbiology and Immunology, INJE University College of Medicine, Busan, Korea.
- 6Exercise Biochemistry Laboratory, Korea National Sport University, Seoul, Korea.
- 7Department of Biomaterials Science, College of Natural Resources & Life Science/Life and Industry Convergence Research Institute, Pusan National University, Miryang, Korea.
- 8College of Veterinary Medicine, Kyungpook National University, Daegu, Korea.
- KMID: 2407430
- DOI: http://doi.org/10.5625/lar.2017.33.2.157
Abstract
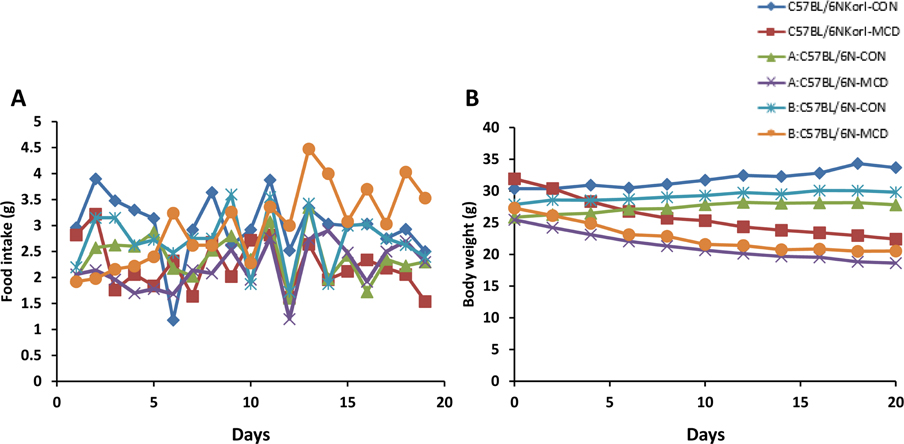
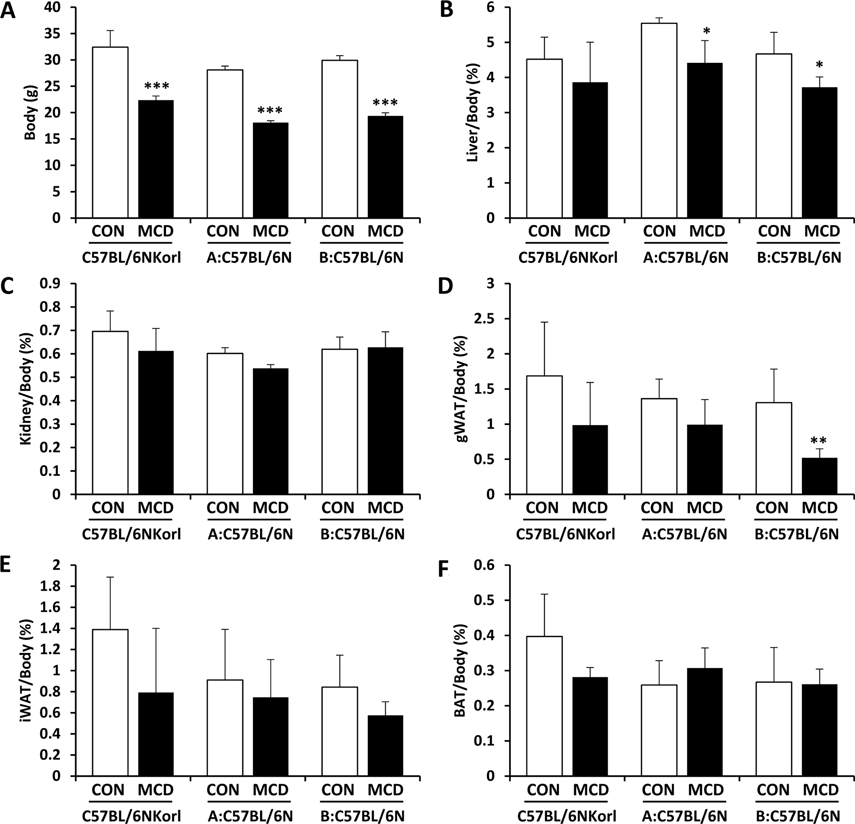
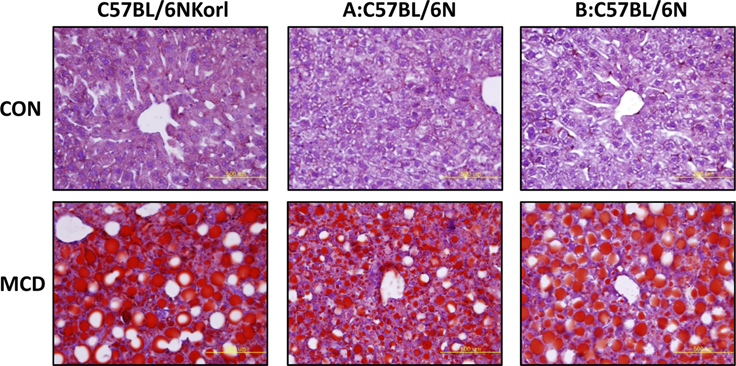
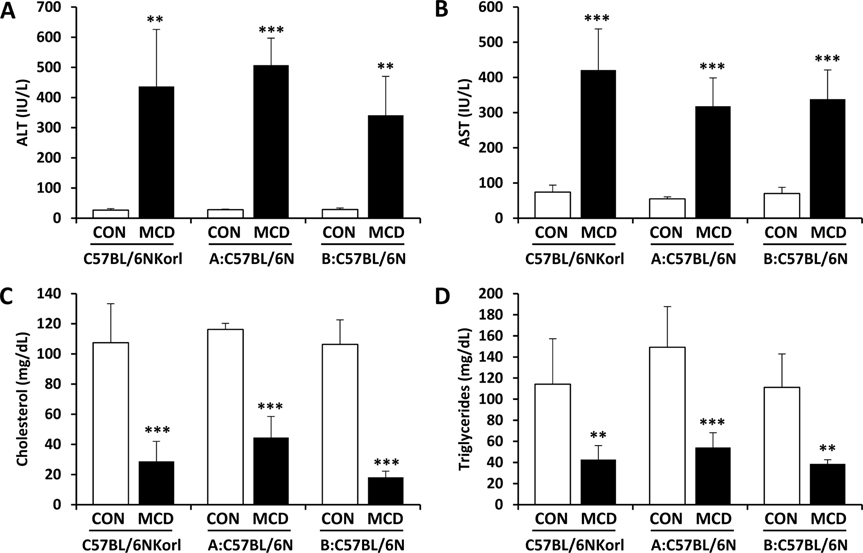
- Non-alcoholic fatty liver disease (NAFLD) is believed to be the most prevalent liver disease worldwide and a major cause of chronic liver injury. It is characterized by lipid accumulation in the absence of significant alcohol consumption and frequently progresses to steatohepatitis, liver fibrosis, and hepatocellular carcinoma. Although many studies have been conducted to better understand NAFLD since it was first recognized, there are still many gaps in knowledge of etiology, prognosis, prevention and treatment. Methionine-choline deficient (MCD) diet, a well-established experimental model of NAFLD in rodents, rapidly and efficiently produces the clinical pathologies including macrovesicular steatosis and leads to disease progression. In this study, we measured the response to MCD diet in C57BL/6N mice obtained from three different sources; Korea NIFDS, USA, and Japan. We evaluated changes in body weight, food consumption, and relative weights of tissues such as liver, kidney, gonadal white adipose tissue, inguinal white adipose tissue, and brown adipose tissue. These basic parameters of mice with an MCD diet were not significantly different among the sources of mice tested. After 3 weeks on an MCD diet, histopathological analyses showed that the MCD diet induced clear fat vacuoles involving most area of the acinus in the liver of all mice. It was accompanied by increased serum activities of alanine aminotransferase and aspartate aminotransferase, and decreased levels of serum triglyceride and cholesterol. In conclusion, the response of C57BL6N mice originating from different sources to the MCD diet showed no significant differences as measured by physiological, biochemical, and histopathological parameters.
MeSH Terms
-
Adipose Tissue, Brown
Adipose Tissue, White
Alanine Transaminase
Alcohol Drinking
Animals
Aspartate Aminotransferases
Body Weight
Carcinoma, Hepatocellular
Cholesterol
Diet
Disease Progression
Fatty Liver*
Gonads
Japan
Kidney
Korea
Liver
Liver Cirrhosis
Liver Diseases
Methionine*
Mice*
Models, Theoretical
Non-alcoholic Fatty Liver Disease
Pathology
Prognosis
Rodentia
Triglycerides
Vacuoles
Weights and Measures
Alanine Transaminase
Aspartate Aminotransferases
Cholesterol
Methionine
Figure
Reference
-
1. Loomba R, Sanyal AJ. The global NAFLD epidemic. Nat Rev Gastroenterol Hepatol. 2013; 10(11):686–690.2. Chalasani N, Younossi Z, Lavine JE, Diehl AM, Brunt EM, Cusi K, Charlton M, Sanyal AJ; American Gastroenterological Association; American Association for the Study of Liver Diseases; American College of Gastroenterologyh. The diagnosis and management of non-alcoholic fatty liver disease: practice guideline by the American Gastroenterological Association, American Association for the Study of Liver Diseases, and American College of Gastroenterology. Gastroenterology. 2012; 142(7):1592–1609.3. Browning JD, Horton JD. Molecular mediators of hepatic steatosis and liver injury. J Clin Invest. 2004; 114(2):147–152.4. Cohen JC, Horton JD, Hobbs HH. Human fatty liver disease: old questions and new insights. Science. 2011; 332(6037):1519–1523.5. Day CP, James OF. Steatohepatitis: a tale of two “hits”? Gastroenterology. 1998; 114(4):842–845.6. Kirsch R, Clarkson V, Shephard EG, Marais DA, Jaffer MA, Woodburne VE, Kirsch RE, Hall Pde L. Rodent nutritional model of non-alcoholic steatohepatitis: species, strain and sex difference studies. J Gastroenterol Hepatol. 2003; 18(11):1272–1282.7. Leclercq IA, Farrell GC, Field J, Bell DR, Gonzalez FJ, Robertson GR. CYP2E1 and CYP4A as microsomal catalysts of lipid peroxides in murine nonalcoholic steatohepatitis. J Clin Invest. 2000; 105(8):1067–1075.8. Bothe GW, Bolivar VJ, Vedder MJ, Geistfeld JG. Genetic and behavioral differences among five inbred mouse strains commonly used in the production of transgenic and knockout mice. Genes Brain Behav. 2004; 3(3):149–157.9. Bryant CD, Zhang NN, Sokoloff G, Fanselow MS, Ennes HS, Palmer AA, McRoberts JA. Behavioral differences among C57BL/6 substrains: implications for transgenic and knockout studies. J Neurogenet. 2008; 22(4):315–331.10. Mulligan MK, Ponomarev I, Boehm SL 2nd, Owen JA, Levin PS, Berman AE, Blednov YA, Crabbe JC, Williams RW, Miles MF, Bergeson SE. Alcohol trait and transcriptional genomic analysis of C57BL/6 substrains. Genes Brain Behav. 2008; 7(6):677–689.11. Mekada K, Abe K, Murakami A, Nakamura S, Nakata H, Moriwaki K, Obata Y, Yoshiki A. Genetic differences among C57BL/6 substrains. Exp Anim. 2009; 58(2):141–149.12. Zurita E, Chagoyen M, Cantero M, Alonso R, González-Neira A, López-Jiménez A, López-Moreno JA, Landel CP, Benítez J, Pazos F, Montoliu L. Genetic polymorphisms among C57BL/6 mouse inbred strains. Transgenic Res. 2011; 20(3):481–489.13. Itagaki H, Shimizu K, Morikawa S, Ogawa K, Ezaki T. Morphological and functional characterization of non-alcoholic fatty liver disease induced by a methionine-choline-deficient diet in C57BL/6 mice. Int J Clin Exp Pathol. 2013; 6(12):2683–2696.14. Machado MV, Cortez-Pinto H. Non-alcoholic fatty liver disease: what the clinician needs to know. World J Gastroenterol. 2014; 20(36):12956–12980.15. Rinella ME, Elias MS, Smolak RR, Fu T, Borensztajn J, Green RM. Mechanisms of hepatic steatosis in mice fed a lipogenic methionine choline-deficient diet. J Lipid Res. 2008; 49(5):1068–1076.16. Yao ZM, Vance DE. The active synthesis of phosphatidylcholine is required for very low density lipoprotein secretion from rat hepatocytes. J Biol Chem. 1988; 263(6):2998–3004.17. Caballero F, Fernández A, Matías N, Martínez L, Fucho R, Elena M, Caballeria J, Morales A, Fernández-Checa JC, García-Ruiz C. Specific contribution of methionine and choline in nutritional nonalcoholic steatohepatitis: impact on mitochondrial S-adenosyl-L-methionine and glutathione. J Biol Chem. 2010; 285(24):18528–18536.18. REITMAN S, FRANKEL S. A colorimetric method for the determination of serum glutamic oxalacetic and glutamic pyruvic transaminases. Am J Clin Pathol. 1957; 28(1):56–63.19. Wei E, Ben Ali Y, Lyon J, Wang H, Nelson R, Dolinsky VW, Dyck JR, Mitchell G, Korbutt GS, Lehner R. Loss of TGH/Ces3 in mice decreases blood lipids, improves glucose tolerance, and increases energy expenditure. Cell Metab. 2010; 11(3):183–193.20. Kim SN, Jung YS, Kwon HJ, Seong JK, Granneman JG, Lee YH. Sex differences in sympathetic innervation and browning of white adipose tissue of mice. Biol Sex Differ. 2016; 7:67.21. Tanaka N, Takahashi S, Fang ZZ, Matsubara T, Krausz KW, Qu A, Gonzalez FJ. Role of white adipose lipolysis in the development of NASH induced by methionine- and cholinedeficient diet. Biochim Biophys Acta. 2014; 1841(11):1596–1607.22. Day CP. Non-alcoholic fatty liver disease: a massive problem. Clin Med (Lond). 2011; 11(2):176–178.23. Ekstedt M, Franzén LE, Mathiesen UL, Thorelius L, Holmqvist M, Bodemar G, Kechagias S. Long-term follow-up of patients with NAFLD and elevated liver enzymes. Hepatology. 2006; 44(4):865–873.24. Sanyal AJ, Brunt EM, Kleiner DE, Kowdley KV, Chalasani N, Lavine JE, Ratziu V, McCullough A. Endpoints and clinical trial design for nonalcoholic steatohepatitis. Hepatology. 2011; 54(1):344–353.25. Takahashi Y, Soejima Y, Fukusato T. Animal models of nonalcoholic fatty liver disease/nonalcoholic steatohepatitis. World J Gastroenterol. 2012; 18(19):2300–2308.26. Sahai A, Malladi P, Melin-Aldana H, Green RM, Whitington PF. Upregulation of osteopontin expression is involved in the development of nonalcoholic steatohepatitis in a dietary murine model. Am J Physiol Gastrointest Liver Physiol. 2004; 287(1):G264–G273.27. Weltman MD, Farrell GC, Liddle C. Increased hepatocyte CYP2E1 expression in a rat nutritional model of hepatic steatosis with inflammation. Gastroenterology. 1996; 111(6):1645–1653.28. London RM, George J. Pathogenesis of NASH: animal models. Clin Liver Dis. 2007; 11(1):55–74. viii29. Yao ZM, Vance DE. Reduction in VLDL, but not HDL, in plasma of rats deficient in choline. Biochem Cell Biol. 1990; 68(2):552–558.30. Rinella ME, Green RM. The methionine-choline deficient dietary model of steatohepatitis does not exhibit insulin resistance. J Hepatol. 2004; 40(1):47–51.31. Machado MV, Michelotti GA, Xie G, Almeida Pereira T, Boursier J, Bohnic B, Guy CD, Diehl AM. Mouse models of diet-induced nonalcoholic steatohepatitis reproduce the heterogeneity of the human disease. PLoS One. 2015; 10(5):e0127991.32. Mashimo T, Serikawa T. Rat resources in biomedical research. Curr Pharm Biotechnol. 2009; 10(2):214–220.33. Croy BA. The 1999 Reginald Thomson Lecture. Custom-built mice: unique discovery tools in biomedical research. Can Vet J. 2000; 41(3):201–206.34. Ardaillou R. Transgenic mice: a major advance in biomedical research. Bull Acad Natl Med. 2009; 193(8):1773–1782.35. Fontaine DA, Davis DB. Attention to Background Strain Is Essential for Metabolic Research: C57BL/6 and the International Knockout Mouse Consortium. Diabetes. 2016; 65(1):25–33.36. Collins S, Martin TL, Surwit RS, Robidoux J. Genetic vulnerability to diet-induced obesity in the C57BL/6J mouse: physiological and molecular characteristics. Physiol Behav. 2004; 81(2):243–248.37. Winzell MS, Ahrén B. The high-fat diet-fed mouse: a model for studying mechanisms and treatment of impaired glucose tolerance and type 2 diabetes. Diabetes. 2004; 53:Suppl 3. S215–S219.38. Sundberg JP, Schofield PN. Commentary: mouse genetic nomenclature. Standardization of strain, gene, and protein symbols. Vet Pathol. 2010; 47(6):1100–1104.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Erratum: Comparative study of fatty liver induced by methionine and choline-deficiency in C57BL/6N mice originating from three different sources
- Compensatory role of C3 convertase on the strain difference for C3 protein expression in FVB/N, C3H/HeN and C57BL/6N mice
- Comparative study of liver injury induced by high-fat methionine- and choline-deficient diet in ICR mice originating from three different sources
- Downregulation of GNAI3 Promotes the Pathogenesis of Methionine/Choline-Deficient Diet-Induced Nonalcoholic Fatty Liver Disease
- Comparative analysis of restraint stress-induced depressive-like phenotypes in C57BL/6N mice derived from three different sources