J Vet Sci.
2017 Dec;18(4):531-540. 10.4142/jvs.2017.18.4.531.
Molecular cloning and characterization of porcine ribosomal protein L21
- Affiliations
-
- 1Biotherapeutics Translational Research Center, Korea Research Institute of Bioscience and Biotechnology, Daejeon 34141, Korea. jwlee@kribb.re.kr
- 2Department of Animal Science and Biotechnology, College of Agriculture and Life Science, Chungnam National University, Daejeon 34134, Korea.
- 3Animal Biotechnology Division, National Institute of Animal Science, Wanju 55365, Korea. hwangss@korea.kr
- KMID: 2398513
- DOI: http://doi.org/10.4142/jvs.2017.18.4.531
Abstract
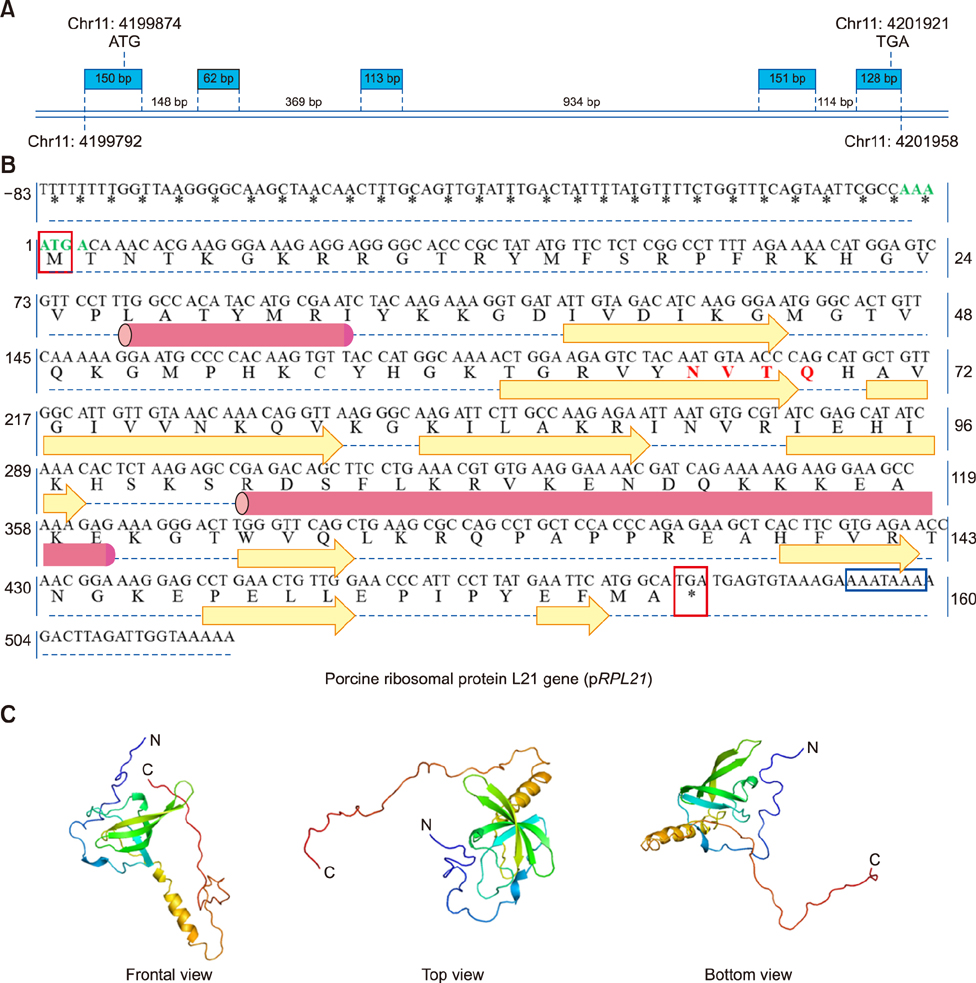
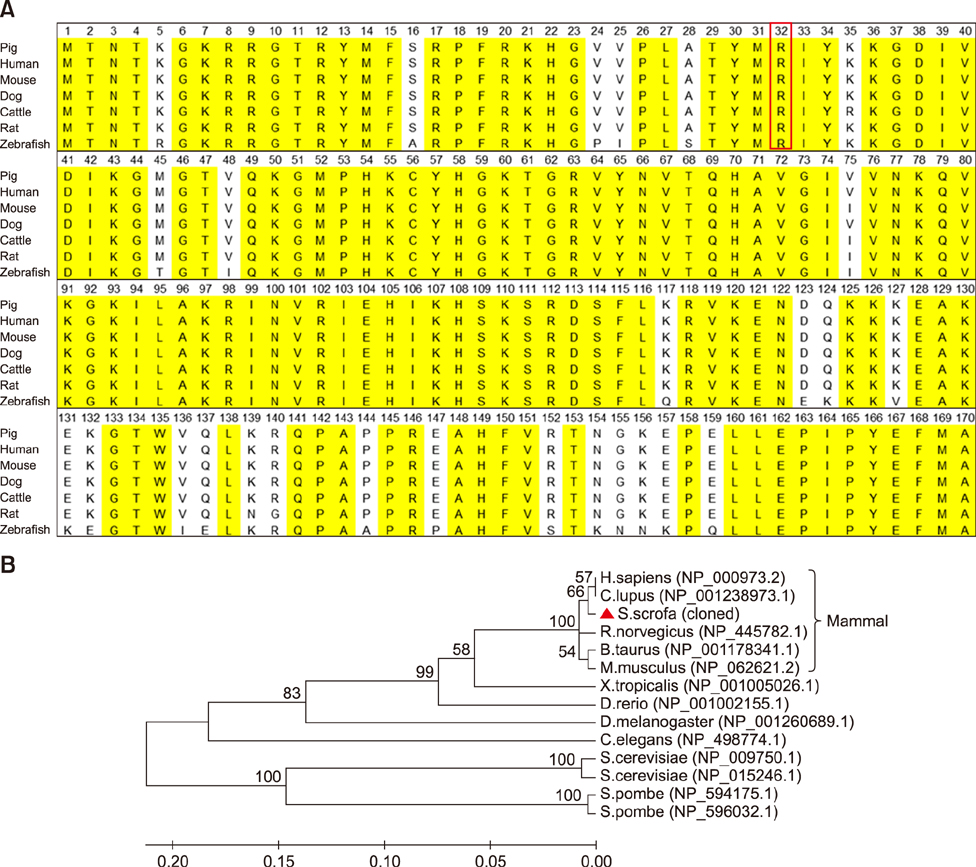
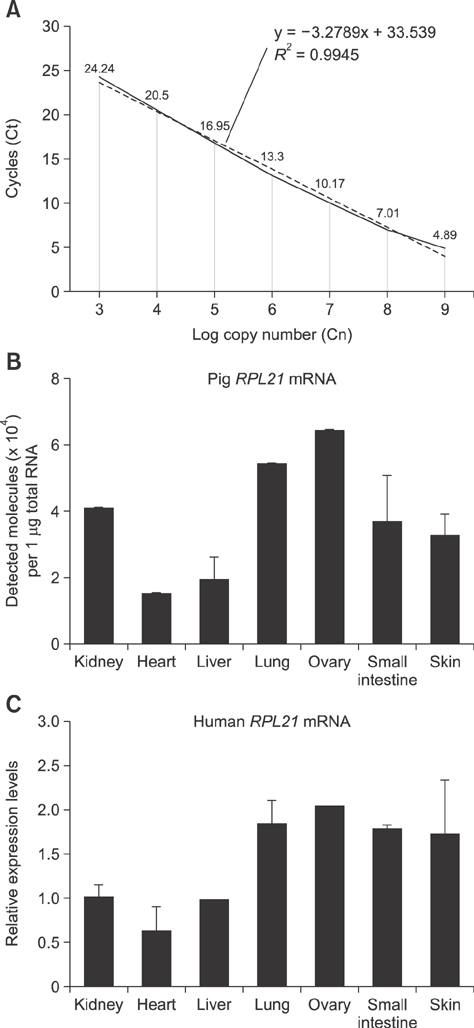
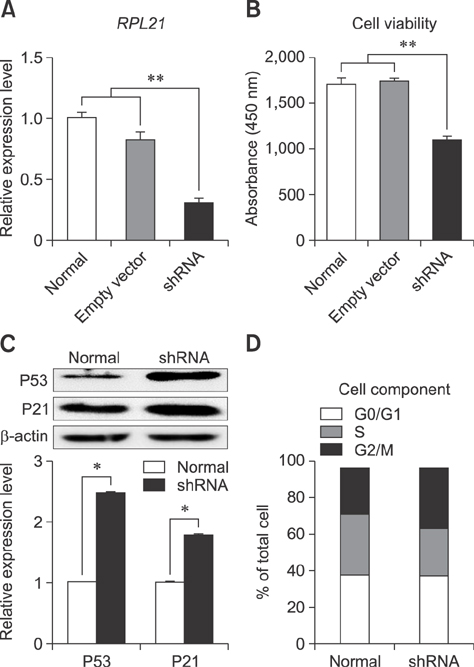
- Ribosomal protein L21 (RPL21) is a structural component of the 60S subunit of the eukaryotic ribosome. This protein has an important role in protein synthesis and the occurrence of hereditary diseases. Pig is a common laboratory model, however, to the best of our knowledge, its RPL21 gene has not been cloned to date. In this study, we cloned and identified the full-length sequence of the pig RPL21 gene for the first time. In addition, we examined its expression pattern and function by using overexpression or knockdown approaches. As a result, we obtained a 604 bp segment that contains a 483 bp open reading frame encoding 160 amino acids. The pig RPL21 gene is located in the "+" strand of chromosome 11, which spans 2167 bp from 4199792 to 4201958. Pig RPL21 protein has nine strands and two helices in its secondary structure. Pig RPL21 is predominantly expressed in ovary and lung, at lower levels in kidney, small intestine, and skin, and at the lowest levels in heart and liver. Furthermore, RPL21 expression is closely connected with cell proliferation and cell cycle arrest. The results are intended to provide useful information for the further study of pig RPL21.
MeSH Terms
Figure
Reference
-
1. Agarwal ML, Agarwal A, Taylor WR, Stark GR. p53 controls both the G2/M and the G1 cell cycle checkpoints and mediates reversible growth arrest in human fibroblasts. Proc Natl Acad Sci U S A. 1995; 92:8493–8497.
Article2. Artimo P, Jonnalagedda M, Arnold K, Baratin D, Csardi G, de Castro E, Duvaud S, Flegel V, Fortier A, Gasteiger E, Grosdidier A, Hernandez C, Ioannidis V, Kuznetsov D, Liechti R, Moretti S, Mostaguir K, Redaschi N, Rossier G, Xenarios I, Stockinger H. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012; 40:W597–W603.
Article3. Bhavsar RB, Makley LN, Tsonis PA. The other lives of ribosomal proteins. Hum Genomics. 2010; 4:327–344.
Article4. Chakraborty A, Uechi T, Kenmochi N. Guarding the ‘translation apparatus’: defective ribosome biogenesis and the p53 signaling pathway. Wiley Interdiscip Rev RNA. 2011; 2:507–522.
Article5. Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003; 31:3497–3500.
Article6. De Angioletti M, Lacerra G, Sabato V, Carestia C. β+45 G → C: a novel silent β-thalassaemia mutation, the first in the Kozak sequence. Br J Haematol. 2004; 124:224–231.
Article7. Deisenroth C, Zhang Y. The ribosomal protein-Mdm2-p53 pathway and energy metabolism: bridging the gap between feast and famine. Genes Cancer. 2011; 2:392–403.
Article8. Devi KR, Chan YL, Wool IG. The primary structure of rat ribosomal protein S4. Biochim Biophys Acta. 1989; 1008:258–262.
Article9. Frigerio JM, Dagorn JC, Iovanna JL. Cloning, sequencing and expression of the L5, L21, L27a, L28, S5, S9, S10 and S29 human ribosomal protein mRNAs. Biochim Biophys Acta. 1995; 1262:64–68.
Article10. Harms J, Schluenzen F, Zarivach R, Bashan A, Gat S, Agmon I, Bartels H, Franceschi F, Yonath A. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell. 2001; 107:679–688.
Article11. Hou YL, Ding X, Hou W, Song B, Wang T, Wang F, Li J, Zhong J, Xu T, Ma BX, Zhu HQ, Li JH, Zhong JC. Overexpression, purification, and pharmacologic evaluation of anticancer activity of ribosomal protein L24 from the giant panda (Ailuropoda melanoleuca). Genet Mol Res. 2013; 12:4735–4750.
Article12. Hsiao LL, Dangond F, Yoshida T, Hong R, Jensen RV, Misra J, Dillon W, Lee KF, Clark KE, Haverty P, Weng Z, Mutter GL, Frosch MP, MacDonald ME, Milford EL, Crum CP, Bueno R, Pratt RE, Mahadevappa M, Warrington JA, Stephanopoulos G, Stephanopoulos G, Gullans SR. A compendium of gene expression in normal human tissues. Physiol Genomics. 2001; 7:97–104.
Article13. Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014; 30:1236–1240.
Article14. Kiefer F, Arnold K, Künzli M, Bordoli L, Schwede T. The SWISS-Model Repository and associated resources. Nucleic Acids Res. 2009; 37:D387–D392.
Article15. Klinge S, Voigts-Hoffmann F, Leibundgut M, Arpagaus S, Ban N. Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science. 2011; 334:941–948.
Article16. Korobeinikova AV, Garber MB, Gongadze GM. Ribosomal proteins: structure, function, and evolution. Biochemistry (Mosc). 2012; 77:562–574.
Article17. Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986; 44:283–292.
Article18. Kwon DJ, Jeon H, Oh KB, Ock SA, Im GS, Lee SS, Im SK, Lee JW, Oh SJ, Park JK, Hwang S. Generation of leukemia inhibitory factor-dependent induced pluripotent stem cells from the Massachusetts General Hospital miniature pig. Biomed Res Int. 2013; 2013:140639.
Article19. Lee C, Kim J, Shin SG, Hwang S. Absolute and relative QPCR quantification of plasmid copy number in Escherichia coli. J Biotechnol. 2006; 123:273–280.
Article20. Lin A, Chan YL, Jones R, Wool IG. The primary structure of rat ribosomal protein S12. The relationship of rat S12 to other ribosomal proteins and a correlation of the amino acid sequences of rat and yeast ribosomal proteins. J Biol Chem. 1987; 262:14343–14351.
Article21. Lindström MS. Emerging functions of ribosomal proteins in gene-specific transcription and translation. Biochem Biophys Res Commun. 2009; 379:167–170.
Article22. Liu JM, Ellis SR. Ribosomes and marrow failure: coincidental association or molecular paradigm. Blood. 2006; 107:4583–4588.
Article23. Loreni F, Francesconi A, Jappelli R, Amaldi F. Analysis of mRNAs under translational control during Xenopus embryogenesis: isolation of new ribosomal protein clones. Nucleic Acids Res. 1992; 20:1859–1863.
Article24. Loreni F, Ruberti I, Bozzoni I, Pierandrei-Amaldi P, Amaldi F. Nucleotide sequence of the L1 ribosomal protein gene of Xenopus laevis: remarkable sequence homology among introns. EMBO J. 1985; 4:3483–3488.
Article25. Nygard AB, Jørgensen CB, Cirera S, Fredholm M. Selection of reference genes for gene expression studies in pig tissues using SYBR green qPCR. BMC Mol Biol. 2007; 8:67.
Article26. Plafker SM, Macara IG. Ribosomal protein L12 uses a distinct nuclear import pathway mediated by importin 11. Mol Cell Biol. 2002; 22:1266–1275.
Article27. Plaks V, Gershon E, Zeisel A, Jacob-Hirsch J, Neeman M, Winterhager E, Rechavi G, Domany E, Dekel N. Blastocyst implantation failure relates to impaired translational machinery gene expression. Reproduction. 2014; 148:87–98.
Article28. Rebouças EL, Costa JJN, Passos MJ, Passos JRS, van den Hurk R, Silva JRV. Real time PCR and importance of housekeepings genes for normalization and quantification of mRNA expression in different tissues. Braz Arch Biol Technol. 2013; 56:143–154.
Article29. Rosendahl G, Andreasen PH, Kristiansen K. Structure and evolution of the Tetrahymena thermophila gene encoding ribosomal protein L21. Gene. 1991; 98:161–167.30. Rosorius O, Fries B, Stauber RH, Hirschmann N, Bevec D, Hauber J. Human ribosomal protein L5 contains defined nuclear localization and export signals. J Biol Chem. 2000; 275:12061–12068.
Article31. Snapp E. Design and use of fluorescent fusion proteins in cell biology. Curr Protoc Cell Biol. 2005; Suppl 27. Unit 21.4.
Article32. Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, Cooke MP, Walker JR, Hogenesch JB. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004; 101:6062–6067.
Article33. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011; 28:2731–2739.
Article34. Taylor WR, Stark GR. Regulation of the G2/M transition by p53. Oncogene. 2001; 20:1803–1815.
Article35. Thorrez L, Van Deun K, Tranchevent LC, Van Lommel L, Engelen K, Marchal K, Moreau Y, Van Mechelen I, Schuit F. Using ribosomal protein genes as reference: a tale of caution. PLoS One. 2008; 3:e1854.
Article36. Wang W, Nag S, Zhang X, Wang MH, Wang H, Zhou J, Zhang R. Ribosomal proteins and human diseases: pathogenesis, molecular mechanisms, and therapeutic implications. Med Res Rev. 2015; 35:225–285.
Article37. Warner JR. Synthesis of ribosomes in Saccharomyces cerevisiae. Microbiol Rev. 1989; 53:256–271.38. Warner JR, McIntosh KB. How common are extraribosomal functions of ribosomal proteins. Mol Cell. 2009; 34:3–11.
Article39. Weisberg RA. Transcription by moonlight: structural basis of an extraribosomal activity of ribosomal protein S10. Mol Cell. 2008; 32:747–748.
Article40. Wool IG. The structure and function of eukaryotic ribosomes. Annu Rev Biochem. 1979; 48:719–754.
Article41. Wool IG, Chan YL, Glück A. Structure and evolution of mammalian ribosomal proteins. Biochem Cell Biol. 1995; 73:933–947.
Article42. Xie M, Kobayashi I, Kiyoshima T, Nagata K, Ookuma Y, Fujiwara H, Sakai H. In situ expression of ribosomal protein L21 in developing tooth germ of the mouse lower first molar. J Mol Histol. 2009; 40:361–367.
Article43. Yusupova G, Yusupov M. High-resolution structure of the eukaryotic 80S ribosome. Annu Rev Biochem. 2014; 83:467–486.
Article44. Zhang W, Hawse J, Huang Q, Sheets N, Miller KM, Horwitz J, Kantorow M. Decreased expression of ribosomal proteins in human age-related cataract. Invest Ophthalmol Vis Sci. 2002; 43:198–204.45. Zhang Y, Lu H. Signaling to p53: ribosomal proteins find their way. Cancer Cell. 2009; 16:369–377.
Article46. Zhou C, Zang D, Jin Y, Wu H, Liu Z, Du J, Zhang J. Mutation in ribosomal protein L21 underlies hereditary hypotrichosis simplex. Hum Mutat. 2011; 32:710–714.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Molecular cloning of ribosomal P protein in Toxoplasma gondii and the availability to detect antibody against recombinant protein in toxoplasmosis patients
- Molecular Cloning of Novel Clathrin Assembly Protein Gene from Rat Brain
- Cloning and Characterization of a Novel Laccase Gene, fvlac7, Based on the Genomic Sequence of Flammulina velutipes
- Characterization of the Recombinant Proteins of Porcine Circovirus Type2 Field Isolate Expressed in the Baculovirus System
- Molecular Cloning and Nucleotide Sequence of the Gene Encoding Gp44 Protein of Suri strain: an Attenuated Classical Swine Fever Virus