Nat Prod Sci.
2017 Sep;23(3):162-168. 10.20307/nps.2017.23.3.162.
Inhibitory Activity of Cordyceps bassiana Extract on LPS-induced Inflammation in RAW 264.7 Cells by Suppressing NF-κB Activation
- Affiliations
-
- 1Department of Biochemistry, Kangwon National University, Chunchon 200-701, Republic of Korea.
- 2College of Pharmacy, Kangwon National University, Chunchon 200-701, Republic of Korea. haeilp@kangwon.ac.kr
- 3Department of Science Education, Kangwon National University, Chunchon 200-701, Republic of Korea.
- 4Department of Food Science and Human Nutrition, Michigan State University, East Lansing, MI48824, USA.
- 5Institute for Bio-Medical Convergence, Catholic Kwandong University, Incheon 404-834, Republic of Korea. ung97330@gmail.com
- KMID: 2393796
- DOI: http://doi.org/10.20307/nps.2017.23.3.162
Abstract
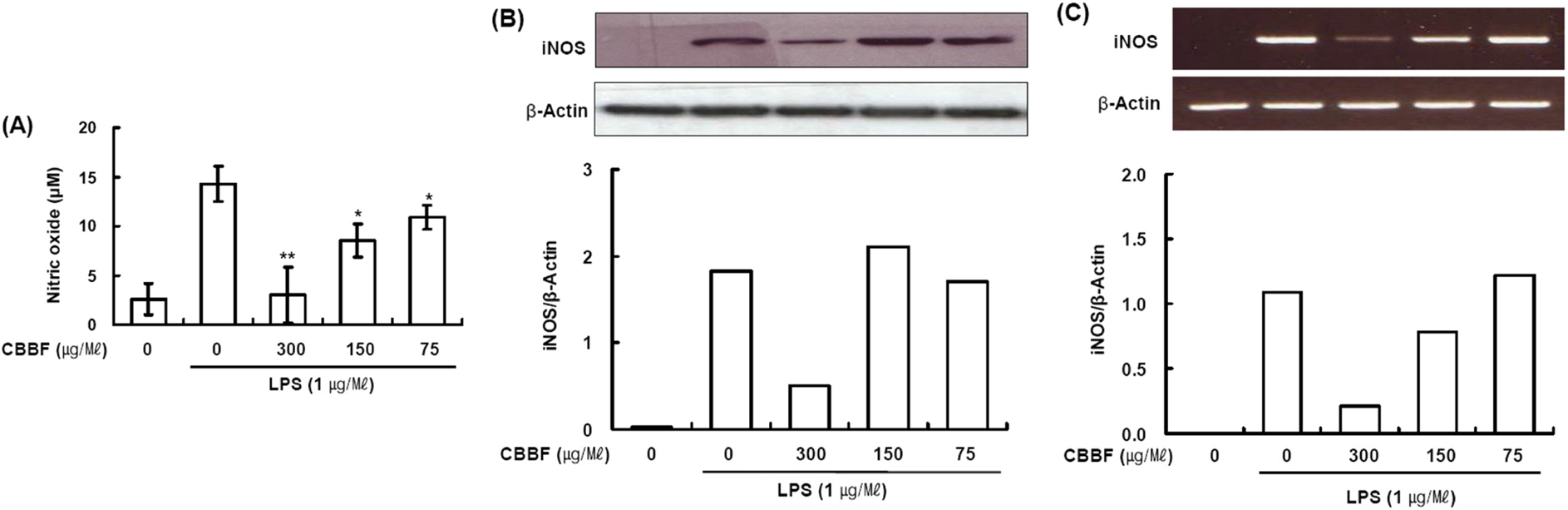
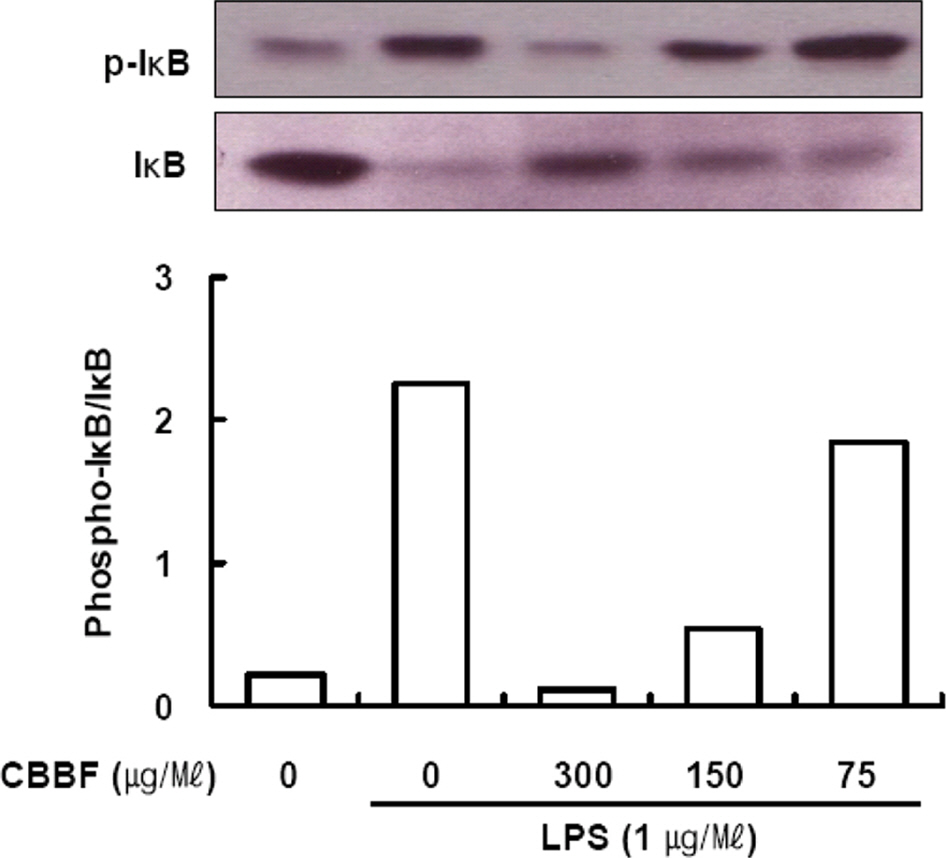
- Cordyceps bassiana has long been used as an oriental medicine and reported to possess diverse biological activities. The fruiting bodies of Cordyceps bassiana was extracted with ethanol and then further fractionated with n-hexane, ethyl acetate, n-butanol and water. The butanol fraction from Cordyceps bassiana (CBBF) exhibited the most effective in anti-inflammatory activity in RAW 264.7 macrophages and the roles of CBBF on the anti-inflammation cascade in LPS-stimulated RAW 264.7 cells were studied. To investigate the mechanism by which CBBF inhibits NO, iNOS and COX-2, the activation of IκB and MAPKs in LPS-activated macrophage were examined. Our present results demonstrated that CBBF inhibits NO production and iNOS expression in LPS-stimulated RAW 264.7 macrophage cells, and these effects were mediated through the inhibition of IκB-α, JNK and p38 phosphorylation. Also, CBBF suppressed activation of MAPKs including p38 and SAPK/JNK. Furthermore, CBBF significantly suppressed LPS-induced intracellular ROS generation. Its inhibition on iNOS expression, together with its antioxidant activity, may support its anti-inflammatory activity. Thus Cordyceps bassiana can be used as a useful medicinal food or drug for further studies.
Keyword
MeSH Terms
Figure
Reference
-
References
(1). Zhou X.., Gong Z.., Su Y.., Lin J.., Tang K. J.Pharm. Pharmacol. 2009. 61:279–291.(2). Sung G. H.., Hywel-Jones N. L.., Sung J. M.., Luangsa-Ard J. J.., Shrestha B.., Spatafora J. W.Stud. Mycol. 2007. 57:5–59.(3). Priest F. G.., Goodfellow M.Applied Microbial Systematics (9eds.); Kluwer Academic Publishers: Dordrecht. P,. 2000. 203–230.(4). Schaeffenberg B. Z.Pflanzenkrankh. Pflanzensch. 1955. 62:544–549.(5). Li Z.., Li C.., Huang B.., Fan M.Chinese Science Bulletin. 2001. 46:751–753.(6). Kim K. M.., Kwon Y. G.., Chung H. T.., Yun Y. G.., Pae H. O.., Han J. A.., Ha K. S.., Kim T. W.., Kim Y. M.Toxicol. Appl. Pharmacol. 2003. 190:1–8.(7). Park Y. M.., Won J. H.., Kim Y. H.., Choi J. W.., Park H. J.., Lee K. T. J.Ethnopharmacol. 2005. 101:120–128.(8). Kim B. C.., Choi J. W.., Hong H. Y.., Lee S. A.., Hong S.., Park E. H.., Kim S. J.., Lim C. J. J.Ethnopharmacol. 2006. 106:364–371.(9). Paterson R. R.Phytochemistry. 2008. 69:1469–1495.(10). Wadsworth T. L.., Koop D. R.Biochem. Pharmacol. 1999. 57:941–949.(11). Makarov S. S.Mol. Med. Today. 2000. 6:441–448.
Article(12). D'Acquisto F.., May M. J.., Ghosh S.Mol. Interv. 2002. 2:22–35.(13). Moynagh P. N. J.Cell Sci. 2005. 118:4589–4592.(14). Giri S.., Rattan R.., Singh A. K.., Singh I. J.Immunol. 2004. 173:5196–5208.(15). Islam S.., Hassan F.., Mu M. M.., Ito H.., Koide N.., Mori I.., Yoshida T.., Yokochi T.Microbiol. Immunol. 2004. 48:729–736.(16). Chan E. D.., Riches D. W.Am. J. Physiol. Cell Physiol. 2001. 280:C441–C450.(17). Hommes D. W.., Peppelenbosch M. P.., van Deventer S. J.Gut. 2003. 52:144–151.(18). Kim S. H.., Johnson V. J.., Shin T. Y.., Sharma R. P.Exp. Biol. Med(Maywood). 2004. 229:203–213.(19). Bai S. K.., Lee S. J.., Na H. J.., Ha K. S.., Han J. A.., Lee H.., Kwon Y. G.., Chung C. K.., Kim Y. M.Exp. Mol. Med. 2005. 37:323–334.(20). Kim J. H.., Kim D. H.., Baek S. H.., Lee H. J.., Kim M. R.., Kwon H. J.., Lee C. H.Biochem. Pharmacol. 2006. 71:1198–1205.(21). Torres M.., Forman H. J.Biofactors. 2003. 17:287–296.(22). Suh S. J.., Chung T. W.., Son M. J.., Kim S. H.., Moon T. C.., Son K. H.., Kim H. P.., Chang H. W.., Kim C. H.Arch. Biochem. Biophys. 2006. 447:136–146.(23). Suh W.., Nam G.., Yang W. S.., Sung G. H.., Shim S. H.., Cho J. Y.Biomol. Ther. 2017. 25:165–170.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Anti-inflammatory activities of Scolopendra subspinipes mutilans in RAW 264.7 cells
- Anti-inflammatory Activity of Standardized Fraction from Inula helenium L. via Suppression of NF-κB Pathway in RAW 264.7 Cells
- Anti-inflammatory effects of proanthocyanidin-rich red rice extract via suppression of MAPK, AP-1 and NF-κB pathways in Raw 264.7 macrophages
- Estragole Exhibits Anti-inflammatory Activity with the Regulation of NF-κB and Nrf-2 Signaling Pathways in LPS-induced RAW 264.7 cells
- Rhodanthpyrone A and B play an anti-inflammatory role by suppressing the nuclear factor-κB pathway in macrophages