Ann Lab Med.
2015 Jan;35(1):118-122. 10.3343/alm.2015.35.1.118.
Alteration of the SETBP1 Gene and Splicing Pathway Genes SF3B1, U2AF1, and SRSF2 in Childhood Acute Myeloid Leukemia
- Affiliations
-
- 1Department of Laboratory Medicine, Chonnam National University Hwasun Hospital, Hwasun, Korea. mgshin@chonnam.ac.kr
- 2Brain Korea 21 Plus Project, Chonnam National University Medical School, Gwangju, Korea.
- 3Laboratory of Metabolism, National Cancer Institute, National Institutes of Health, Bethesda, MD, USA.
- 4Department of Pediatrics, Chonnam National University Hwasun Hospital, Hwasun, Korea.
- 5Environmental Health Center for Childhood Leukemia and Cancer, Chonnam National University Hwasun Hospital, Hwasun, Korea.
- KMID: 2363158
- DOI: http://doi.org/10.3343/alm.2015.35.1.118
Abstract
- BACKGROUND
Recurrent somatic SET-binding protein 1 (SETBP1) and splicing pathway gene mutations have recently been found in atypical chronic myeloid leukemia and other hematologic malignancies. These mutations have been comprehensively analyzed in adult AML, but not in childhood AML. We investigated possible alteration of the SETBP1, splicing factor 3B subunit 1 (SF3B1), U2 small nuclear RNA auxiliary factor 1 (U2AF1), and serine/arginine-rich splicing factor 2 (SRSF2) genes in childhood AML.
METHODS
Cytogenetic and molecular analyses were performed to reveal chromosomal and genetic alterations. Sequence alterations in the SETBP1, SF3B1, U2AF1, and SRSF2 genes were examined by using direct sequencing in a cohort of 53 childhood AML patients.
RESULTS
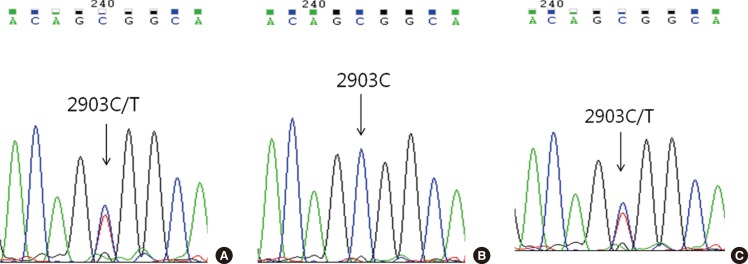
Childhood AML patients did not harbor any recurrent SETBP1 gene mutations, although our study did identify a synonymous mutation in one patient. None of the previously reported aberrations in the mutational hotspot of SF3B1, U2AF1, and SRSF2 were identified in any of the 53 patients.
CONCLUSIONS
Alterations of the SETBP1 gene or SF3B1, U2AF1, and SRSF2 genes are not common genetic events in childhood AML, implying that the mutations are unlikely to exert a driver effect in myeloid leukemogenesis during childhood.
MeSH Terms
-
Adolescent
Carrier Proteins/*genetics
Child
Child, Preschool
Cohort Studies
Cytogenetic Analysis
DNA Mutational Analysis
Female
Gene Frequency
Genotype
Humans
Infant
Leukemia, Myeloid, Acute/*genetics/pathology
Male
Nuclear Proteins/*genetics
Phosphoproteins/*genetics
Polymorphism, Single Nucleotide
RNA Splicing
Ribonucleoprotein, U2 Small Nuclear/*genetics
Ribonucleoproteins/*genetics
Carrier Proteins
Nuclear Proteins
Phosphoproteins
Ribonucleoprotein, U2 Small Nuclear
Ribonucleoproteins
Figure
Reference
-
1. Hoischen A, van Bon BW, Gilissen C, Arts P, van Lier B, Steehouwer M, et al. De novo mutations of SETBP1 cause Schinzel-Giedion syndrome. Nat Genet. 2010; 42:483–485. PMID: 20436468.
Article2. Piazza R, Valletta S, Winkelmann N, Redaelli S, Spinelli R, Pirola A, et al. Recurrent SETBP1 mutations in atypical chronic myeloid leukemia. Nat Genet. 2013; 45:18–24. PMID: 23222956.
Article3. Damm F, Itzykson R, Kosmider O, Droin N, Renneville A, Chesnais V, et al. SETBP1 mutations in 658 patients with myelodysplastic syndromes, chronic myelomonocytic leukemia and secondary acute myeloid leukemias. Leukemia. 2013; 27:1401–1403. PMID: 23443343.
Article4. Minakuchi M, Kakazu N, Gorrin-Rivas MJ, Abe T, Copeland TD, Ueda K, et al. Identification and characterization of SEB, a novel protein that binds to the acute undifferentiated leukemia-associated protein SET. Eur J Biochem. 2001; 268:1340–1351. PMID: 11231286.
Article5. Cristóbal I, Blanco FJ, Garcia-Orti L, Marcotegui N, Vicente C, Rifon J, et al. SETBP1 overexpression is a novel leukemogenic mechanism that predicts adverse outcome in elderly patients with acute myeloid leukemia. Blood. 2010; 115:615–625. PMID: 19965692.
Article6. Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011; 478:64–69. PMID: 21909114.
Article7. Thol F, Suchanek KJ, Koenecke C, Stadler M, Platzbecker U, Thiede C, et al. SETBP1 mutation analysis in 944 patients with MDS and AML. Leukemia. 2013; 27:2072–2075. PMID: 23648668.
Article8. Creutzig U, van den, Gibson B, Dworzak MN, Adachi S, de Bont E, et al. Diagnosis and management of acute myeloid leukemia in children and adolescents: recommendations from an international expert panel. Blood. 2012; 120:3187–3205. PMID: 22879540.
Article9. Makishima H, Visconte V, Sakaguchi H, Jankowska AM, Abu Kar S, Jerez A, et al. Mutations in the spliceosome machinery, a novel and ubiquitous pathway in leukemogenesis. Blood. 2012; 119:3203–3210. PMID: 22323480.
Article10. Choi HJ, Kim HR, Shin MG, Kook H, Kim HJ, Shin JH, et al. Spectra of chromosomal aberrations in 325 leukemia patients and implications for the development of new molecular detection systems. J Korean Med Sci. 2011; 26:886–892. PMID: 21738341.
Article11. Murati A, Brecqueville M, Devillier R, Mozziconacci MJ, Gelsi-Boyer V, Birnbaum D. Myeloid malignancies: mutations, models and management. BMC Cancer. 2012; 12:304. PMID: 22823977.
Article12. Liang DC, Liu HC, Yang CP, Jaing TH, Hung IJ, Yeh TC, et al. Cooperating gene mutations in childhood acute myeloid leukemia with special reference on mutations of ASXL1, TET2, IDH1, IDH2, and DNMT3A. Blood. 2013; 121:2988–2995. PMID: 23365461.
Article13. Shiba N, Ohki K, Park MJ, Sotomatsu M, Kudo K, Ito E, et al. SETBP1 mutations in juvenile myelomonocytic leukaemia and myelodysplastic syndrome but not in paediatric acute myeloid leukaemia. Br J Haematol. 2014; 164:142–159. PMID: 24033149.14. Je EM, Yoo NJ, Kim YJ, Kim MS, Lee SH. Mutational analysis of splicing machinery genes SF3B1, U2AF1 and SRSF2 in myelodysplasia and other common tumors. Int J Cancer. 2013; 133:260–265. PMID: 23280334.15. Kar SA, Jankowska A, Makishima H, Visconte V, Jerez A, Sugimoto Y, et al. Spliceosomal gene mutations are frequent events in the diverse mutational spectrum of chronic myelomonocytic leukemia but largely absent in juvenile myelomonocytic leukemia. Haematologica. 2013; 98:107–113. PMID: 22773603.
Article16. Hunt R, Sauna ZE, Ambudkar SV, Gottesman MM, Kimchi-Sarfaty C. Silent (synonymous) SNPs: should we care about them? Methods Mol Biol. 2009; 578:23–39. PMID: 19768585.
Article17. Chamary JV, Parmley JL, Hurst LD. Hearing silence: non-neutral evolution at synonymous sites in mammals. Nat Rev Genet. 2006; 7:98–108. PMID: 16418745.
Article18. Ho PA, Kuhn J, Gerbing RB, Pollard JA, Zeng R, Miller KL, et al. WT1 synonymous single nucleotide polymorphism rs16754 correlates with higher mRNA expression and predicts significantly improved outcome in favorable-riskpediatric acute myeloid leukemia: a report from the children's oncology group. J Clin Oncol. 2011; 29:704–711. PMID: 21189390.19. Damm F, Heuser M, Morgan M, Yun H, Grosshennig A, Göhring G, et al. Single nucleotide polymorphism in the mutational hotspot of WT1 predicts a favorable outcome in patients with cytogenetically normal acute myeloid leukemia. J Clin Oncol. 2010; 28:578–585. PMID: 20038731.
Article20. Ho PA, Kopecky KJ, Alonzo TA, Gerbing RB, Miller KL, Kuhn J, et al. Prognostic implications of the IDH1 synonymous SNP rs11554137 in pediatric and adult AML: a report from the Children's Oncology Group and SWOG. Blood. 2011; 118:4561–4566. PMID: 21873548.
Article21. National Center for Biotechnology Information, U.S. National Library of Medicine. SNP database. Updated on Oct 2013. http://www.ncbi.nlm.nih.gov/SNP/index.html.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- What is new in acute myeloid leukemia classification?
- Mutational Analysis of CDKN2 (p16-INK4A/MTS1) Gene in Childhood Acute Leukemia
- Treatments for children and adolescents with AML
- Informative Gene Selection Method in Tumor Classification
- A Case of CD7+, CD4-, CD8-, CD3-acute T cell lymphoblastic leukemia