J Korean Med Sci.
2015 Dec;30(12):1784-1792. 10.3346/jkms.2015.30.12.1784.
Urinary Nucleic Acid TSPAN13-to-S100A9 Ratio as a Diagnostic Marker in Prostate Cancer
- Affiliations
-
- 1Department of Urology, College of Medicine, Chungbuk National University, Cheongju, Korea. wjkim@chungbuk.ac.kr
- 2Bio-Medical Science Co. Ltd, Daejeon, Korea.
- 3Nucleic Acid Research Center, Inc., Daejeon Oriental Hospital of Daejeon University, Daejeon, Korea.
- 4Department of Biochemistry, Dong-Eui University, Busan, Korea.
- 5Department of Food and Biotechnology, Chung-Ang University, Seoul, Korea.
- KMID: 2359962
- DOI: http://doi.org/10.3346/jkms.2015.30.12.1784
Abstract
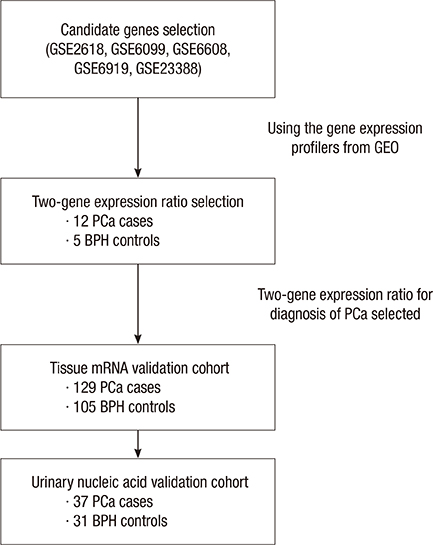
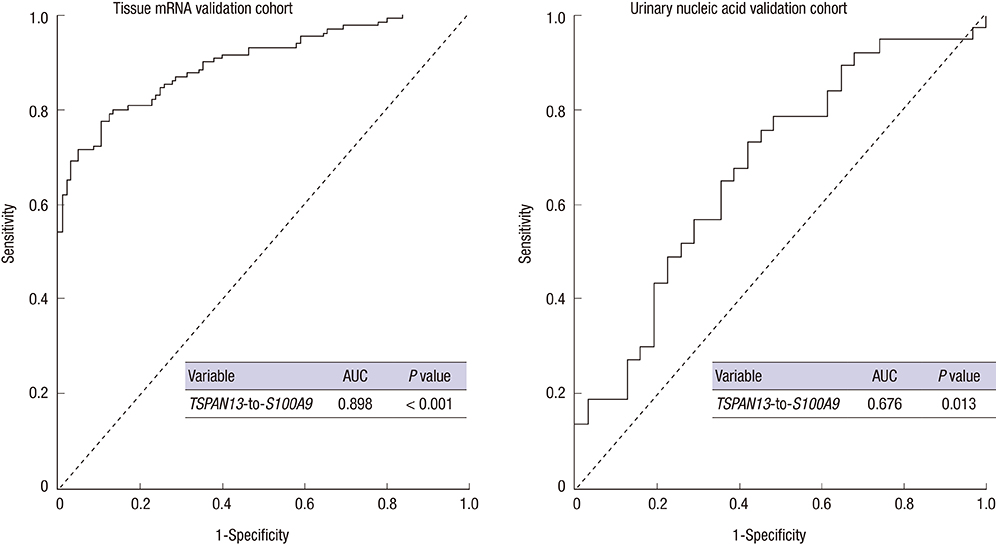
- The potential use of urinary nucleic acids as diagnostic markers in prostate cancer (PCa) was evaluated. Ninety-five urine samples and 234 prostate tissue samples from patients with PCa and benign prostatic hyperplasia (BPH) were analyzed. Micro-array analysis was used to identify candidate genes, which were verified by the two-gene expression ratio and validated in tissue mRNA and urinary nucleic acid cohorts. Real-time quantitative polymerase chain reaction (qPCR) was used to measure urinary nucleic acid levels and tissue mRNA expression. The TSPAN13-to-S100A9 ratio was selected to determine the diagnostic value of urinary nucleic acids in PCa (P = 0.037) and shown to be significantly higher in PCa than in BPH in the mRNA and nucleic acid cohort analyses (P < 0.001 and P = 0.013, respectively). Receiver operating characteristic (ROC) analysis showed that the area under the ROC curve was 0.898 and 0.676 in tissue mRNA cohort and urinary nucleic acid cohort, respectively. The TSPAN13-to-S100A9 ratio showed a strong potential as a diagnostic marker for PCa. The present results suggest that the analysis of urine supernatant can be used as a simple diagnostic method for PCa that can be adapted to the clinical setting in the future.
Keyword
MeSH Terms
-
Aged
Aged, 80 and over
Biomarkers, Tumor/*genetics/*urine
Calgranulin B/*genetics
Cohort Studies
Humans
Male
Middle Aged
Nucleic Acids/*genetics/*urine
Oligonucleotide Array Sequence Analysis
Prostate/metabolism
Prostatic Hyperplasia/diagnosis/genetics/urine
Prostatic Neoplasms/diagnosis/*genetics/*urine
RNA, Messenger/genetics/metabolism
RNA, Neoplasm/genetics/metabolism
ROC Curve
Real-Time Polymerase Chain Reaction
Tetraspanins/*genetics
Biomarkers, Tumor
Calgranulin B
Nucleic Acids
RNA, Messenger
RNA, Neoplasm
Tetraspanins
Figure
Reference
-
1. Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012; 62:10–29.2. Jung KW, Park S, Kong HJ, Won YJ, Boo YK, Shin HR, Park EC, Lee JS. Cancer statistics in Korea: incidence, mortality and survival in 2006-2007. J Korean Med Sci. 2010; 25:1113–1121.3. Jung KW, Yim SH, Kong HJ, Hwang SY, Won YJ, Lee JK, Shin HR. Cancer survival in Korea 1993-2002: a population-based study. J Korean Med Sci. 2007; 22:S5–S10.4. Roobol MJ, Zappa M, Määttänen L, Ciatto S. The value of different screening tests in predicting prostate biopsy outcome in screening for prostate cancer data from a multicenter study (ERSPC). Prostate. 2007; 67:439–446.5. Ralla B, Stephan C, Meller S, Dietrich D, Kristiansen G, Jung K. Nucleic acid-based biomarkers in body fluids of patients with urologic malignancies. Crit Rev Clin Lab Sci. 2014; 51:200–231.6. Gahan PB, Swaminathan R. Circulating nucleic acids in plasma and serum. Recent developments. Ann N Y Acad Sci. 2008; 1137:1–6.7. Casadio V, Calistri D, Salvi S, Gunelli R, Carretta E, Amadori D, Silvestrini R, Zoli W. Urine cell-free DNA integrity as a marker for early prostate cancer diagnosis: a pilot study. Biomed Res Int. 2013; 2013:270457.8. De Maio G, Rengucci C, Zoli W, Calistri D. Circulating and stool nucleic acid analysis for colorectal cancer diagnosis. World J Gastroenterol. 2014; 20:957–967.9. Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD, Knippers R. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001; 61:1659–1665.10. Yun SJ, Yan C, Jeong P, Kang HW, Kim YH, Kim EA, Lee OJ, Kim WT, Moon SK, Kim IY, et al. Comparison of mRNA, protein, and urinary nucleic acid levels of S100A8 and S100A9 between prostate cancer and BPH. Ann Surg Oncol. 2015; 22:2439–2445.11. Ma XJ, Wang Z, Ryan PD, Isakoff SJ, Barmettler A, Fuller A, Muir B, Mohapatra G, Salunga R, Tuggle JT, et al. A two-gene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer Cell. 2004; 5:607–616.12. Tomlins SA, Mehra R, Rhodes DR, Shah RB, Rubin MA, Bruening E, Makarov V, Chinnaiyan AM. Whole transcriptome amplification for gene expression profiling and development of molecular archives. Neoplasia. 2006; 8:153–162.13. Tomlins SA, Mehra R, Rhodes DR, Cao X, Wang L, Dhanasekaran SM, Kalyana-Sundaram S, Wei JT, Rubin MA, Pienta KJ, et al. Integrative molecular concept modeling of prostate cancer progression. Nat Genet. 2007; 39:41–51.14. Chandran UR, Ma C, Dhir R, Bisceglia M, Lyons-Weiler M, Liang W, Michalopoulos G, Becich M, Monzon FA. Gene expression profiles of prostate cancer reveal involvement of multiple molecular pathways in the metastatic process. BMC Cancer. 2007; 7:64.15. Kim YJ, Yoon HY, Kim SK, Kim YW, Kim EJ, Kim IY, Kim WJ. EFEMP1 as a novel DNA methylation marker for prostate cancer: array-based DNA methylation and expression profiling. Clin Cancer Res. 2011; 17:4523–4530.16. Mead R, Duku M, Bhandari P, Cree IA. Circulating tumour markers can define patients with normal colons, benign polyps, and cancers. Br J Cancer. 2011; 105:239–245.17. Yu J, Zhu T, Wang Z, Zhang H, Qian Z, Xu H, Gao B, Wang W, Gu L, Meng J, et al. A novel set of DNA methylation markers in urine sediments for sensitive/specific detection of bladder cancer. Clin Cancer Res. 2007; 13:7296–7304.18. Miyake M, Sugano K, Sugino H, Imai K, Matsumoto E, Maeda K, Fukuzono S, Ichikawa H, Kawashima K, Hirabayashi K, et al. Fibroblast growth factor receptor 3 mutation in voided urine is a useful diagnostic marker and significant indicator of tumor recurrence in non-muscle invasive bladder cancer. Cancer Sci. 2010; 101:250–258.19. Vinci S, Giannarini G, Selli C, Kuncova J, Villari D, Valent F, Orlando C. Quantitative methylation analysis of BCL2, hTERT, and DAPK promoters in urine sediment for the detection of non-muscle-invasive urothelial carcinoma of the bladder: a prospective, two-center validation study. Urol Oncol. 2011; 29:150–156.20. Zancan M, Galdi F, Di Tonno F, Mazzariol C, Orlando C, Malentacchi F, Agostini M, Maran M, Del Bianco P, Fabricio AS, et al. Evaluation of cell-free DNA in urine as a marker for bladder cancer diagnosis. Int J Biol Markers. 2009; 24:147–155.21. Chang HW, Tsui KH, Shen LC, Huang HW, Wang SN, Chang PL. Urinary cell-free DNA as a potential tumor marker for bladder cancer. Int J Biol Markers. 2007; 22:287–294.22. Szarvas T, Kovalszky I, Bedi K, Szendroi A, Majoros A, Riesz P, Füle T, László V, Kiss A, Romics I. Deletion analysis of tumor and urinary DNA to detect bladder cancer: urine supernatant versus urine sediment. Oncol Rep. 2007; 18:405–409.23. Zancan M, Franceschini R, Mimmo C, Vianello M, Di Tonno F, Mazzariol C, Malossini G, Gion M. Free DNA in urine: a new marker for bladder cancer? Preliminary data. Int J Biol Markers. 2005; 20:134–136.24. Casadio V, Calistri D, Tebaldi M, Bravaccini S, Gunelli R, Martorana G, Bertaccini A, Serra L, Scarpi E, Amadori D, et al. Urine cell-free DNA integrity as a marker for early bladder cancer diagnosis: preliminary data. Urol Oncol. 2013; 31:1744–1750.25. Reid JF, Lusa L, De Cecco L, Coradini D, Veneroni S, Daidone MG, Gariboldi M, Pierotti MA. Limits of predictive models using microarray data for breast cancer clinical treatment outcome. J Natl Cancer Inst. 2005; 97:927–930.26. Jansen MP, Sieuwerts AM, Look MP, Ritstier K, Meijer-van Gelder ME, van Staveren IL, Klijn JG, Foekens JA, Berns EM. HOXB13-to-IL17BR expression ratio is related with tumor aggressiveness and response to tamoxifen of recurrent breast cancer: a retrospective study. J Clin Oncol. 2007; 25:662–668.27. Gordon GJ, Jensen RV, Hsiao LL, Gullans SR, Blumenstock JE, Ramaswamy S, Richards WG, Sugarbaker DJ, Bueno R. Translation of microarray data into clinically relevant cancer diagnostic tests using gene expression ratios in lung cancer and mesothelioma. Cancer Res. 2002; 62:4963–4967.28. De Rienzo A, Dong L, Yeap BY, Jensen RV, Richards WG, Gordon GJ, Sugarbaker DJ, Bueno R. Fine-needle aspiration biopsies for gene expression ratio-based diagnostic and prognostic tests in malignant pleural mesothelioma. Clin Cancer Res. 2011; 17:310–316.29. Zimmer DB, Cornwall EH, Landar A, Song W. The S100 protein family: history, function, and expression. Brain Res Bull. 1995; 37:417–429.30. Donato R. S100: a multigenic family of calcium-modulated proteins of the EF-hand type with intracellular and extracellular functional roles. Int J Biochem Cell Biol. 2001; 33:637–668.31. Ehrchen JM, Sunderkötter C, Foell D, Vogl T, Roth J. The endogenous Toll-like receptor 4 agonist S100A8/S100A9 (calprotectin) as innate amplifier of infection, autoimmunity, and cancer. J Leukoc Biol. 2009; 86:557–566.32. Gebhardt C, Németh J, Angel P, Hess J. S100A8 and S100A9 in inflammation and cancer. Biochem Pharmacol. 2006; 72:1622–1631.33. Hermani A, Hess J, De Servi B, Medunjanin S, Grobholz R, Trojan L, Angel P, Mayer D. Calcium-binding proteins S100A8 and S100A9 as novel diagnostic markers in human prostate cancer. Clin Cancer Res. 2005; 11:5146–5152.