Lab Anim Res.
2013 Sep;29(3):162-167. 10.5625/lar.2013.29.3.162.
Proteomic analysis of liver in miniature pigs according to developmental stages using two-dimensional electrophoresis and matrix-assisted laser desorption/ionization-time of flight mass spectrometry
- Affiliations
-
- 1Department of Biomedical Laboratory Science, College of Biomedical Sciences, Soonchunhyang University, Asan, Korea.
- 2Department of Pathology and Laboratory of Immune Regulation, College of Medicine, Seoul National University, Seoul, Korea.
- 3Laboratory of Developmental Biology and Genetics, College of Veterinary Medicine, Seoul National University, Seoul, Korea. snumouse@snu.ac.kr
- 4Transitional Research Institute, College of Medicine, Seoul National University, Seoul, Korea.
- 5Biomedical Center for Animal Resource and Development, Bio-Max Institute, Seoul National University, Seoul, Korea.
- 6Department of Biochemistry, Hanyang University, Ansan, Korea.
- 7Interdisciplinary Program for Bioinformatics, Program for Cancer Biology and Bio-Max Institute, Seoul National University, Seoul, Korea.
- KMID: 2312109
- DOI: http://doi.org/10.5625/lar.2013.29.3.162
Abstract
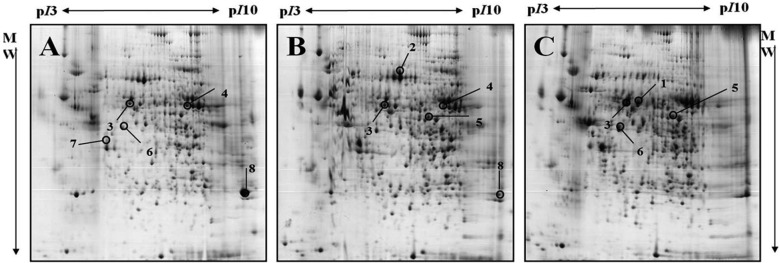
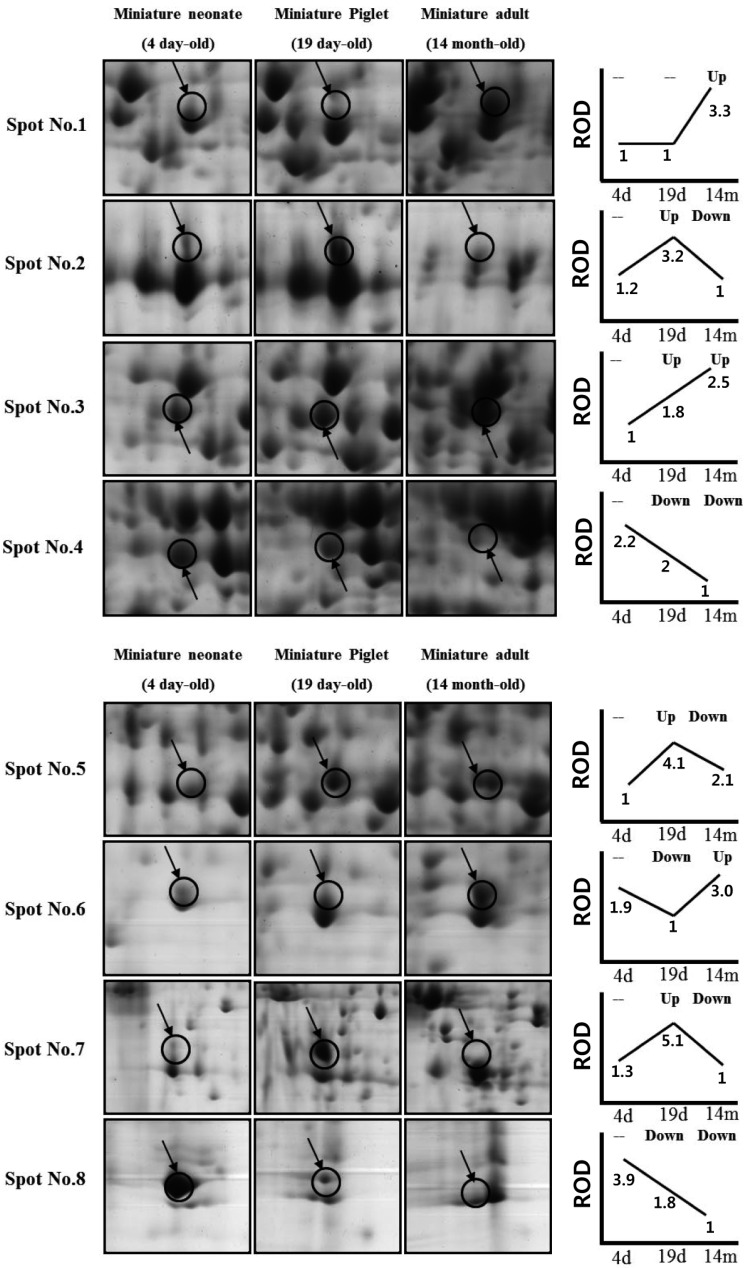
- Due to the shortage of human organ donors for transplant, various studies of xenotransplantation, or the use of animal organs instead of human organs, have been carried out. The organs of porcine are thought to be safer and of a more suitable size for xenotransplantationthan those of nonhuman primates. Understanding the levels of expression of proteins, and their post-translational regulation, would be very practical between different species and among developing stages, though the molecular profiling for xenotransplantation has been rarely studied for porcine, while that of human and rodent is well known. Here, in this present study, we report protein regulation of the developing stages of liver (4-day old neonate, 19-day old piglet and 14-month old adult miniature pigs) using 2-DE and MALDI-TOF. From images of the three different stages, a total of 8 spotswhich were differently regulated were identified, and 5 spots were identified with MALDI-TOF MS. The data presented within this study provides critical direction relating to the development of livers of miniature pigs, which will assist future proteome analysis of the liver, and advance our understanding of the hurdles facing xenotransplantaion.
Keyword
MeSH Terms
Figure
Reference
-
1. Junghans P, Kaehne T, Beyer M, Metges CC, Schwerin M. Dietary protein-related changes in hepatic transcription correspond to modifications in hepatic protein expression in growing pigs. J Nutr. 2004; 134(1):43–47. PMID: 14704291.
Article2. Pan TL, Goto S, Lord R, Huang YC, Huang CM, Wang PW, Lin YC, Kawamoto S, Ono K, Liao PC, Lin CL, Lai CY, Chang HL, Lan CH, Lee TH, Wang YC, Wu ML, Jawan B, Cheng YF, Chen ST, Chen CL. Proteome analysis in liver transplantation. Transplant Proc. 2001; 33(1-2):156. PMID: 11266756.
Article3. Platt JL. Physiologic barriers to xenotransplantation. Transplant Proc. 2000; 32(7):1547–1548. PMID: 11119828.
Article4. Cooper DK, Gollackner B, Sachs DH. Will the pig solve the transplantation backlog? Annu Rev Med. 2002; 53:133–147. PMID: 11818467.
Article5. Tucker A, Belcher C, Moloo B, Bell J, Mazzulli T, Humar A, Hughes A, McArdle P, Talbot A. The production of transgenic pigs for potential use in clinical xenotransplantation: baseline clinical pathology and organ size studies. Xenotransplantation. 2002; 9(3):203–208. PMID: 11983018.
Article6. Steiner S, Anderson NL. Pharmaceutical proteomics. Ann N Y Acad Sci. 2000; 919:48–51. PMID: 11083096.
Article7. Görg A, Obermaier C, Boguth G, Harder A, Scheibe B, Wildgruber R, Weiss W. The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis. 2000; 21(6):1037–1053. PMID: 10786879.
Article8. Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996; 68(5):850–858. PMID: 8779443.9. Alnouti Y, Klaassen CD. Tissue distribution, ontogeny, and regulation of aldehyde dehydrogenase (Aldh) enzymes mRNA by prototypical microsomal enzyme inducers in mice. Toxicol Sci. 2008; 101(1):51–64. PMID: 17998271.
Article10. Akram M, Ali Shah SM, Asif HM, Shaheen G, Shamim T, Khan MI, Ullah A, Ahmed K. Comparative study of similarity and identity of human albumin with some selected organism albumin. J Med Plants Res. 2011; 5(19):4974–4976.11. Magin TM, Jorcano JL, Franke WW. Cytokeratin expression in simple epithelia. II. cDNA cloning and sequence characteristics of bovine cytokeratin A (no. 8). Differentiation. 1986; 30(3):254–264. PMID: 2422084.12. Parker KA, Bruzik JP, Steitz JA. An in vitro interaction between the human U3 snRNP and 28S rRNA sequences near the alpha-sarcin site. Nucleic Acids Res. 1988; 16(22):10493–10509. PMID: 2974535.13. Chen J, Yu L, Li D, Gao Q, Wang J, Huang X, Bi G, Wu H, Zhao S. Human CRYL1, a novel enzyme-crystallin overexpressed in liver and kidney and downregulated in 58% of liver cancer tissues from 60 Chinese patients, and four new homologs from other mammalians. Gene. 2003; 302(1-2):103–113. PMID: 12527201.
Article14. Chen FC, Chuang TJ. ESTviewer: a web interface for visualizing mouse, rat, cattle, pig and chicken conserved ESTs in human genes and human alternatively spliced variants. Bioinformatics. 2005; 21(10):2510–2513. PMID: 15722373.
Article15. Moller M, Berg F, Riquet J, Pomp D, Archibald A, Anderson S, Feve K, Zhang Y, Rothschild M, Milan D, Andersson L, Tuggle CK. High-resolution comparative mapping of pig Chromosome 4, emphasizing the FAT1 region. Mamm Genome. 2004; 15(9):717–731. PMID: 15389320.
Article16. Kahlem P, Dörken B, Schmitt CA. Cellular senescence in cancer treatment: friend or foe? J Clin Invest. 2004; 113(2):169–174. PMID: 14722606.
Article17. Besada V, Diaz M, Becker M, Ramos Y, Castellanos-Serra L, Fichtner I. Proteomics of xenografted human breast cancer indicates novel targets related to tamoxifen resistance. Proteomics. 2006; 6(3):1038–1048. PMID: 16385476.
Article18. Coggin JH Jr, Barsoum AL, Rohrer JW, Thurnher M, Zeis M. Contemporary definitions of tumor specific antigens, immunogens and markers as related to the adaptive responses of the cancer-bearing host. Anticancer Res. 2005; 25(3c):2345–2355. PMID: 16080461.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Proteomic analysis of pancreas in miniature pigs according to developmental stages using two-dimensional electrophoresis and matrix-assisted laser desorption/ionization-time of flight mass spectrometry
- Identification of a Marker Protein for Cardiac Ischemia and Reperfusion Injury by Two-Dimensional Gel Electrophoresis and Matrix-Assisted Laser Desorption Ionization Mass Spectrometry
- MALDI-TOF-MS Fingerprinting Provides Evidence of Urosepsis caused by Aerococcus urinae
- Risk of inaccurate species identification by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and of false carbapenem resistance by automated susceptibility analysis of Enterobacter spp.
- Proteomic analysis of domestic pig pancreas during development using two-dimensional electrophoresis and matrix-assisted laser desorption/ionization-time of flight mass spectrometry