Korean J Hematol.
2007 Mar;42(1):33-42. 10.5045/kjh.2007.42.1.33.
Differentially Expressed Cellular Gene Profiles between Healthy HIV-infected Koreans and AIDS Patients
- Affiliations
-
- 1Division of AIDS, Center for Immunology and Pathology, National Institute of Health, Seoul, Korea. jooshil@nih.go.kr
- KMID: 2252266
- DOI: http://doi.org/10.5045/kjh.2007.42.1.33
Abstract
-
BACKGROUND: The global effect of HIV infection on the host cell gene expression profiles in healthy HIV-infected patients, as long-term non-progressors, remains largely unknown. To identify the cellular genes related with HIV infection and delayed disease progression in vivo, the host gene expression profiles between healthy HIV-infected Koreans and AIDS patients were investigated.
METHODS
Differential expression gene analysis was performed via oligonucleotide microarray with using Magic-oligo 10K chip. Ten HIV-uninfected persons and 10 HIV-infected patients (healthy HIV-infected patients vs. AIDS patients. respectively) were studied.
RESULTS
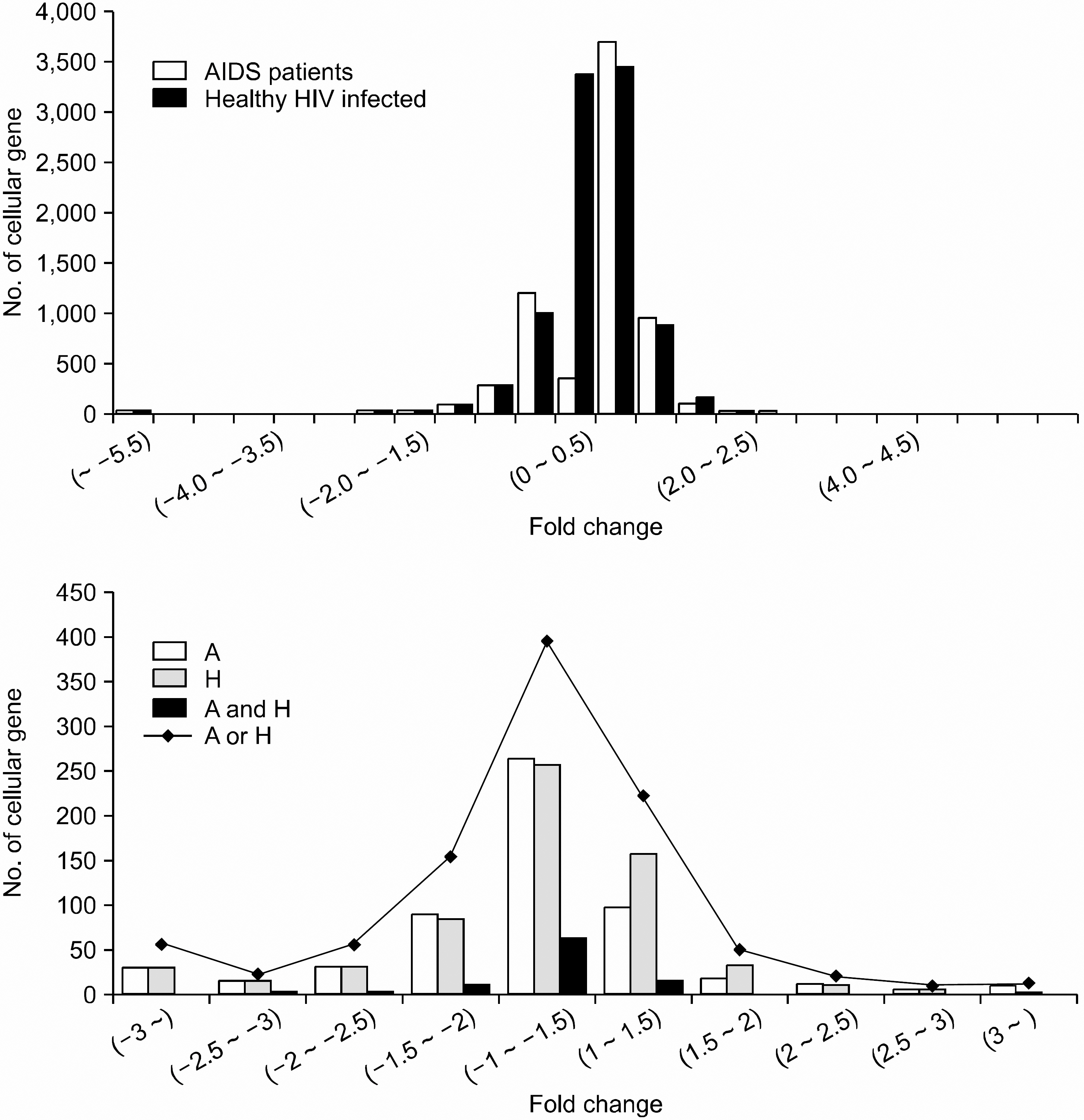
Only 10.8% (1,097 genes) of the total genes, that is, 331 up-regulated genes and 766 down- regulated genes were differentially expressed with more than a two-fold change in the HIV-infected persons as compared to those of the HIV-uninfected persons. Especially, 97 genes (8.8%) among 1,097 genes were commonly up- or down-regulated in both the healthy HIV-infected patients and the AIDS patients. 187 genes were differently expressed on the gene expression analysis between the healthy HIV-infected patients and the AIDS patients. Twenty-eight genes out of them showed very significant differences with a P value <0.01. Especially, tripartite motif (TRIM) 14 protein and interferon gamma receptor 2 were dramatically up-regulated in healthy HIV-infected patients, while death-associated protein, DNA directed RNA polymerase II polypeptide A and STAT were over-expressed in AIDS patients.
CONCLUSION
Although this microarray study has some limitations, the above results will be helpful for performing more detailed, future functional studies on the differentially expressed genes related to HIV infection and delayed disease progression in vivo.
Keyword
MeSH Terms
Figure
Reference
-
1). Geiss GK., Bumgarner RE., An MC, et al. Large-scale monitoring of host cell gene expression during HIV-1 infection using cDNA microarrays. Virology. 2000. 266:8–16.
Article2). Swingler S., Mann A., Jacque J, et al. HIV-1 Nef mediates lymphocyte chemotaxis and activation by infected macrophages. Nat Med. 1999. 5:997–1003.
Article3). Izmailova E., Bertley FM., Huang Q, et al. HIV-1 Tat reprograms immature dendritic cells to express chemoattractants for activated T cells and macrophages. Nat Med. 2003. 9:191–7.
Article4). de la Fuente C., Santiago F., Deng L, et al. Gene expression profile of HIV-1 Tat expressing cells: a close interplay between proliferative and differentiation signals. BMC Biochem. 2002. 3:14.5). Chun TW., Fauci AS. Latent reservoirs of HIV: obstacles to the eradication of virus. Proc Natl Acad Sci U S A. 1999. 96:10958–61.
Article6). Chun TW., Justement JS., Lempicki RA, et al. Gene expression and viral prodution in latently infected, resting CD4+ T cells in viremic versus aviremic HIV-infected individuals. Proc Natl Acad Sci U S A. 2003. 100:1908–13.
Article7). Almeida CA., Price P., French MA. Immune activation in patients infected with HIV type 1 and maintaining suppression of viral replication by highly active antiretroviral therapy. AIDS Res Hum Retroviruses. 2002. 18:1351–5.8). Wilson SA., Sieiro-Vazquez C., Edwards NJ, et al. Cloning and characterization of hIF2, a human homologue of bacterial translation initiation factor 2, and its interaction with HIV-1 matrix. Biochem J. 1999. 342(Pt 1):97–103.
Article9). Endo-Munoz L., Warby T., Harrich D., McMillan NA. Phosphorylation of HIV Tat by PKR increases interaction with TAR RNA and enhances transcription. Virol J. 2005. 2:17.10). Chen W., Tang Z., Fortina P, et al. Ethanol potentiates HIV-1 gp120-induced apoptosis in human neurons via both the death receptor and NMDA receptor pathways. Virology. 2005. 334:59–73.
Article11). Cimarelli A., Luban J. Translation elongation factor 1-alpha interacts specifically with the human immunodeficiency virus type 1 Gag polyprotein. J Virol. 1999. 73:5388–401.
Article12). Perales C., Carrasco L., Ventoso I. Cleavage of eIF4G by HIV-1 protease: effects on translation. FEBS Lett. 2003. 533:89–94.
Article13). Szabo J., Cervenak L., Toth FD, et al. Soluble gC1q-R/p33, a cell protein that binds to the globular “heads” of C1q, effectively inhibits the growth of HIV-1 strains in cell cultures. Clin Immunol. 2001. 99:222–31.14). van't Wout AB., Lehrman GK., Mikheeva SA, et al. Cellular gene expression upon human immunodeficiency virus type 1 infection of CD4(+)-T-cell lines. J Virol. 2003. 77:1392–402.15). Shaheduzzaman S., Krishnan V., Petrovic A, et al. Effects of HIV-1 Nef on cellular gene expression profiles. J Biomed Sci. 2002. 9:82–96.
Article16). Jowett JB., Planelles V., Poon B., Shah NP., Chen ML., Chen IS. The human immunodeficiency virus type 1 vpr gene arrests infected T cells in the G2 + M phase of the cell cycle. J Virol. 1995. 69:6304–13.
Article17). Rey O., Canon J., Krogstad P. HIV-1 Gag protein associates with F-actin present in microfilaments. Virology. 1996. 220:530–4.
Article18). Kawai T., Matsumoto M., Takeda K., Sanjo H., Akira S. ZIP kinase, a novel serine/threonine kinase which mediates apoptosis. Mol Cell Biol. 1998. 18:1642–51.
Article19). Chun RF., Jeang KT. Requirements for RNA polymerase II carboxyl-terminal domain for activated transcription of human retroviruses human T-cell lymphotropic virus I and HIV-1. J Biol Chem. 1996. 271:27888–94.
Article20). Miller ED., Smith JA., Lichtinger M., Wang L., Su L. Activation of the signal transducer and activator of transcription 1 signaling pathway in thymocytes from HIV-1-infected human thymus. AIDS. 2003. 17:1269–77.
Article21). Hottiger MO., Nabel GJ. Interaction of human immunodeficiency virus type 1 Tat with the transcriptional coactivators p300 and CREB binding protein. J Virol. 1998. 72:8252–6.
Article22). Smith VP., Alcami A. Inhibition of interferons by ectromelia virus. J Virol. 2002. 76:1124–34.
Article23). Song B., Javanbakht H., Perron M., Park DH., Strem-lau M., Sodroski J. Retrovirus restriction by TRIM5-alpha variants from Old World and New World primates. J Virol. 2005. 79:3930–7.24). Sedger LM., Shows DM., Blanton RA, et al. IFN-gamma mediates a novel antiviral activity through dynamic modulation of TRAIL and TRAIL receptor expression. J Immunol. 1999. 163:920–6.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Clinical Guidelines for the Treatment and Prevention of Opportunistic Infections in HIV-infected Koreans
- Clinical Guidelines for the Diagnosis and Treatment of HIV/AIDS in HIV-infected Koreans
- The 2013 Clinical Guidelines for the Diagnosis and Treatment of HIV/AIDS in HIV-Infected Koreans
- The 2015 Clinical Guidelines for the Diagnosis and Treatment of HIV/AIDS in HIV-Infected Koreans
- Summary of 2021 Clinical Guidelines for the Diagnosis and Treatment of HIV/AIDS in HIV-infected Koreans