Anat Cell Biol.
2011 Mar;44(1):69-78. 10.5115/acb.2011.44.1.69.
Identification of genes concordantly expressed with Atoh1 during inner ear development
- Affiliations
-
- 1Department of Anatomy, Brain Korea 21 Project for Medical Science, College of Medicine, Yonsei University, Seoul, Korea. bokj@yuhs.ac
- KMID: 2168870
- DOI: http://doi.org/10.5115/acb.2011.44.1.69
Abstract
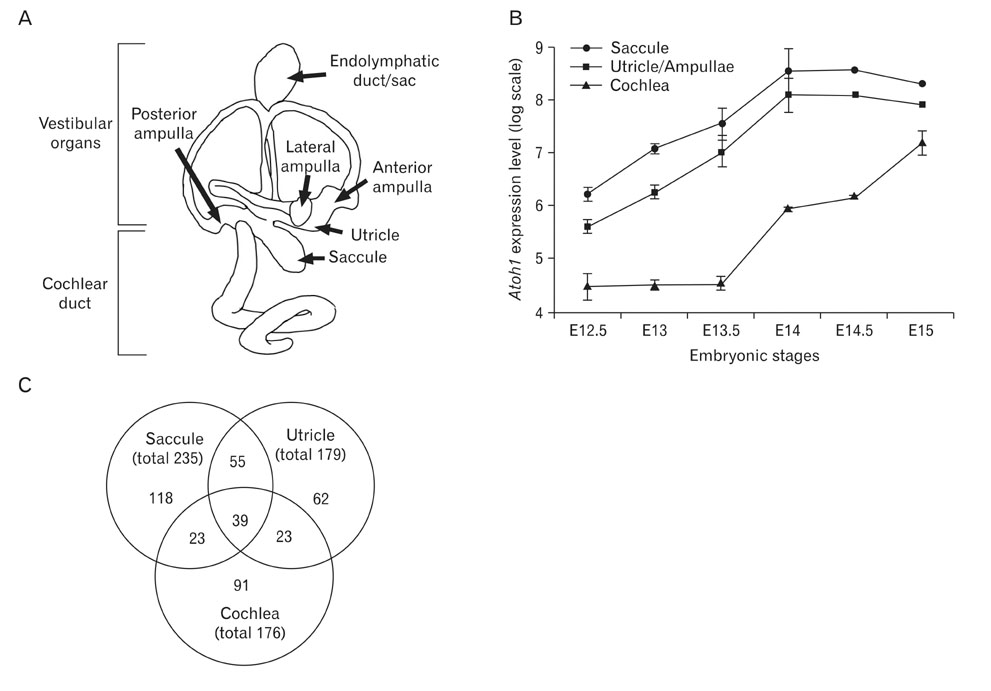
- The inner ear is composed of a cochlear duct and five vestibular organs in which mechanosensory hair cells play critical roles in receiving and relaying sound and balance signals to the brain. To identify novel genes associated with hair cell differentiation or function, we analyzed an archived gene expression dataset from embryonic mouse inner ear tissues. Since atonal homolog 1a (Atoh1) is a well known factor required for hair cell differentiation, we searched for genes expressed in a similar pattern with Atoh1 during inner ear development. The list from our analysis includes many genes previously reported to be involved in hair cell differentiation such as Myo6, Tecta, Myo7a, Cdh23, Atp6v1b1, and Gfi1. In addition, we identified many other genes that have not been associated with hair cell differentiation, including Tekt2, Spag6, Smpx, Lmod1, Myh7b, Kif9, Ttyh1, Scn11a and Cnga2. We examined expression patterns of some of the newly identified genes using real-time polymerase chain reaction and in situ hybridization. For example, Smpx and Tekt2, which are regulators for cytoskeletal dynamics, were shown specifically expressed in the hair cells, suggesting a possible role in hair cell differentiation or function. Here, by reanalyzing archived genetic profiling data, we identified a list of novel genes possibly involved in hair cell differentiation.
Keyword
MeSH Terms
Figure
Reference
-
1. Dror AA, Avraham KB. Hearing loss: mechanisms revealed by genetics and cell biology. Annu Rev Genet. 2009. 43:411–437.2. Li H, Liu H, Heller S. Pluripotent stem cells from the adult mouse inner ear. Nat Med. 2003. 9:1293–1299.3. Izumikawa M, Minoda R, Kawamoto K, Abrashkin KA, Swiderski DL, Dolan DF, Brough DE, Raphael Y. Auditory hair cell replacement and hearing improvement by Atoh1 gene therapy in deaf mammals. Nat Med. 2005. 11:271–276.4. Oshima K, Shin K, Diensthuber M, Peng AW, Ricci AJ, Heller S. Mechanosensitive hair cell-like cells from embryonic and induced pluripotent stem cells. Cell. 2010. 141:704–716.5. Kelley MW. Regulation of cell fate in the sensory epithelia of the inner ear. Nat Rev Neurosci. 2006. 7:837–849.6. Bermingham NA, Hassan BA, Price SD, Vollrath MA, Ben-Arie N, Eatock RA, Bellen HJ, Lysakowski A, Zoghbi HY. Math1: an essential gene for the generation of inner ear hair cells. Science. 1999. 284:1837–1841.7. Woods C, Montcouquiol M, Kelley MW. Math1 regulates development of the sensory epithelium in the mammalian cochlea. Nat Neurosci. 2004. 7:1310–1318.8. Gubbels SP, Woessner DW, Mitchell JC, Ricci AJ, Brigande JV. Functional auditory hair cells produced in the mammalian cochlea by in utero gene transfer. Nature. 2008. 455:537–541.9. Sajan SA, Warchol ME, Lovett M. Toward a systems biology of mouse inner ear organogenesis: gene expression pathways, patterns and network analysis. Genetics. 2007. 177:631–653.10. Wallis D, Hamblen M, Zhou Y, Venken KJ, Schumacher A, Grimes HL, Zoghbi HY, Orkin SH, Bellen HJ. The zinc finger transcription factor Gfi1, implicated in lymphomagenesis, is required for inner ear hair cell differentiation and survival. Development. 2003. 130:221–232.11. Hertzano R, Montcouquiol M, Rashi-Elkeles S, Elkon R, Yücel R, Frankel WN, Rechavi G, Möröy T, Friedman TB, Kelley MW, Avraham KB. Transcription profiling of inner ears from Pou4f3(ddl/ddl) identifies Gfi1 as a target of the Pou4f3 deafness gene. Hum Mol Genet. 2004. 13:2143–2153.12. Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001. 98:5116–5121.13. Morsli H, Choo D, Ryan A, Johnson R, Wu DK. Development of the mouse inner ear and origin of its sensory organs. J Neurosci. 1998. 18:3327–3335.14. Chen P, Johnson JE, Zoghbi HY, Segil N. The role of Math1 in inner ear development: uncoupling the establishment of the sensory primordium from hair cell fate determination. Development. 2002. 129:2495–2505.15. Lumpkin EA, Collisson T, Parab P, Omer-Abdalla A, Haeberle H, Chen P, Doetzlhofer A, White P, Groves A, Segil N, Johnson JE. Math1-driven GFP expression in the developing nervous system of transgenic mice. Gene Expr Patterns. 2003. 3:389–395.16. Mustapha M, Weil D, Chardenoux S, Elias S, El-Zir E, Beckmann JS, Loiselet J, Petit C. An alpha-tectorin gene defect causes a newly identified autosomal recessive form of sensorineural pre-lingual non-syndromic deafness, DFNB21. Hum Mol Genet. 1999. 8:409–412.17. Scott HS, Kudoh J, Wattenhofer M, Shibuya K, Berry A, Chrast R, Guipponi M, Wang J, Kawasaki K, Asakawa S, Minoshima S, Younus F, Mehdi SQ, Radhakrishna U, Papasavvas MP, Gehrig C, Rossier C, Korostishevsky M, Gal A, Shimizu N, Bonne-Tamir B, Antonarakis SE. Insertion of beta-satellite repeats identifies a transmembrane protease causing both congenital and childhood onset autosomal recessive deafness. Nat Genet. 2001. 27:59–63.18. Ahmed ZM, Morell RJ, Riazuddin S, Gropman A, Shaukat S, Ahmad MM, Mohiddin SA, Fananapazir L, Caruso RC, Husnain T, Khan SN, Riazuddin S, Griffith AJ, Friedman TB, Wilcox ER. Mutations of MYO6 are associated with recessive deafness, DFNB37. Am J Hum Genet. 2003. 72:1315–1322.19. Everett LA, Morsli H, Wu DK, Green ED. Expression pattern of the mouse ortholog of the Pendred's syndrome gene (Pds) suggests a key role for pendrin in the inner ear. Proc Natl Acad Sci U S A. 1999. 96:9727–9732.20. Stover EH, Borthwick KJ, Bavalia C, Eady N, Fritz DM, Rungroj N, Giersch AB, Morton CC, Axon PR, Akil I, Al-Sabban EA, Baguley DM, Bianca S, Bakkaloglu A, Bircan Z, Chauveau D, Clermont MJ, Guala A, Hulton SA, Kroes H, Li Volti G, Mir S, Mocan H, Nayir A, Ozen S, Rodriguez Soriano J, Sanjad SA, Tasic V, Taylor CM, Topaloglu R, Smith AN, Karet FE. Novel ATP6V1B1 and ATP6V0A4 mutations in autosomal recessive distal renal tubular acidosis with new evidence for hearing loss. J Med Genet. 2002. 39:796–803.21. Iguchi N, Tanaka H, Fujii T, Tamura K, Kaneko Y, Nojima H, Nishimune Y. Molecular cloning of haploid germ cell-specific tektin cDNA and analysis of the protein in mouse testis. FEBS Lett. 1999. 456:315–321.22. Tanaka H, Iguchi N, Toyama Y, Kitamura K, Takahashi T, Kaseda K, Maekawa M, Nishimune Y. Mice deficient in the axonemal protein Tektin-t exhibit male infertility and immotile-cilium syndrome due to impaired inner arm dynein function. Mol Cell Biol. 2004. 24:7958–7964.23. Zhang Z, Jones BH, Tang W, Moss SB, Wei Z, Ho C, Pollack M, Horowitz E, Bennett J, Baker ME, Strauss JF 3rd. Dissecting the axoneme interactome: the mammalian orthologue of Chlamydomonas PF6 interacts with sperm-associated antigen 6, the mammalian orthologue of Chlamydomonas PF16. Mol Cell Proteomics. 2005. 4:914–923.24. Conley CA, Fritz-Six KL, Almenar-Queralt A, Fowler VM. Leiomodins: larger members of the tropomodulin (Tmod) gene family. Genomics. 2001. 73:127–139.25. Kemp TJ, Sadusky TJ, Simon M, Brown R, Eastwood M, Sassoon DA, Coulton GR. Identification of a novel stretch-responsive skeletal muscle gene (Smpx). Genomics. 2001. 72:260–271.26. Piddini E, Schmid JA, de Martin R, Dotti CG. The Ras-like GTPase Gem is involved in cell shape remodelling and interacts with the novel kinesin-like protein KIF9. EMBO J. 2001. 20:4076–4087.27. Schindeler A, Lavulo L, Harvey RP. Muscle costameric protein, Chisel/Smpx, associates with focal adhesion complexes and modulates cell spreading in vitro via a Rac1/p38 pathway. Exp Cell Res. 2005. 307:367–380.28. van Rooij E, Quiat D, Johnson BA, Sutherland LB, Qi X, Richardson JA, Kelm RJ Jr, Olson EN. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev Cell. 2009. 17:662–673.29. Trudeau MC, Zagotta WN. Calcium/calmodulin modulation of olfactory and rod cyclic nucleotide-gated ion channels. J Biol Chem. 2003. 278:18705–18708.30. Priest BT, Murphy BA, Lindia JA, Diaz C, Abbadie C, Ritter AM, Liberator P, Iyer LM, Kash SF, Kohler MG, Kaczorowski GJ, MacIntyre DE, Martin WJ. Contribution of the tetrodotoxin-resistant voltage-gated sodium channel NaV1.9 to sensory transmission and nociceptive behavior. Proc Natl Acad Sci U S A. 2005. 102:9382–9387.31. Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006. 126:1121–1133.32. Suzuki M. The Drosophila tweety family: molecular candidates for large-conductance Ca2+-activated Cl- channels. Exp Physiol. 2006. 91:141–147.33. Bordon A, Bosco N, Du Roure C, Bartholdy B, Kohler H, Matthias G, Rolink AG, Matthias P. Enforced expression of the transcriptional coactivator OBF1 impairs B cell differentiation at the earliest stage of development. PLoS One. 2008. 3:e4007.34. Belyantseva IA, Boger ET, Friedman TB. Myosin XVa localizes to the tips of inner ear sensory cell stereocilia and is essential for staircase formation of the hair bundle. Proc Natl Acad Sci U S A. 2003. 100:13958–13963.35. Bok J. Building the mammalian cochlea: an overview. Genes Genomics. 2010. 32:1–7.36. Sobkowicz HM, Slapnick SM, August BK. The kinocilium of auditory hair cells and evidence for its morphogenetic role during the regeneration of stereocilia and cuticular plates. J Neurocytol. 1995. 24:633–653.37. Shimasaki S, Yamamoto E, Murayama E, Kurio H, Kaneko T, Shibata Y, Inai T, Iida H. Subcellular localization of Tektin2 in rat sperm flagellum. Zoolog Sci. 2010. 27:755–761.38. Holzmann D, Ott PM, Felix H. Diagnostic approach to primary ciliary dyskinesia: a review. Eur J Pediatr. 2000. 159:95–98.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Atoh1 as a Coordinator of Sensory Hair Cell Development and Regeneration in the Cochlea
- Identification of multiple hub genes in acute kidney injury after kidney transplantation by bioinformatics analysis
- Identification of differentially expressed genes using an annealing control primer system in periodontitis
- Identification of Up-Regulated Genes in Malignant Glioma with Subtraction Hybridization: Preliminary Screening Studies
- A network-biology approach for identification of key genes and pathways involved in malignant peritoneal mesothelioma