J Pathol Transl Med.
2015 Jul;49(4):310-317. 10.4132/jptm.2015.05.13.
Analysis of Histologic Features Suspecting Anaplastic Lymphoma Kinase (ALK)-Expressing Pulmonary Adenocarcinoma
- Affiliations
-
- 1Department of Pathology, Soonchunhyang University Seoul Hospital, Soonchunhyang University College of Medicine, Seoul, Korea.
- 2Department of Pathology, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea. hanjho@skku.edu
- 3Department of Pathology, Gwangmyeong Sungae Hospital, Gwangmyeong, Korea.
- KMID: 2151140
- DOI: http://doi.org/10.4132/jptm.2015.05.13
Abstract
- BACKGROUND
Since 2007 when anaplastic lymphoma kinase (ALK) rearrangements were discovered in non-small cell lung cancer, the ALK gene has received attention due to ALK-targeted therapy, and a notable treatment advantage has been observed in patients harboring the EML4/ALK translocation. However, using ALK-fluorescence in situ hybridization (FISH) as the standard method has demerits such as high cost, a time-consuming process, dependency on interpretation skill, and tissue preparation. We analyzed the histologic findings which could complement the limitation of ALK-FISH test for pulmonary adenocarcinoma.
METHODS
Two hundred five cases of ALK-positive and 101 of ALK-negative pulmonary adenocarcinoma from January 2007 to May 2013 were enrolled in this study. The histologic findings and ALK immunohistochemistry results were reviewed and compared with the results of ALK-FISH and EGFR/KRAS mutation status.
RESULTS
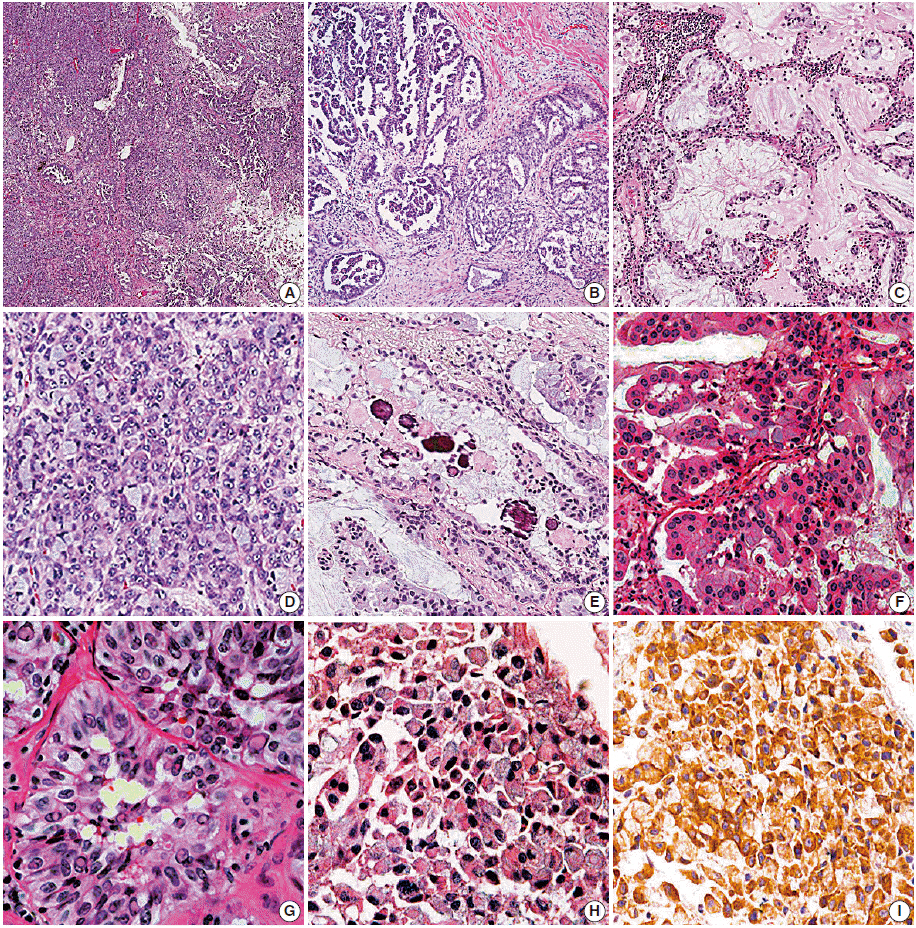
Acinar, cribriform, and solid growth patterns, extracellular and intracellular mucin production, and presence of signet-ring-cell element, and psammoma body were significantly more often present in ALK-positive cancer. In addition, the presence of goblet cell-like cells and presence of nuclear inclusion and groove resembling papillary thyroid carcinoma were common in the ALK-positive group.
CONCLUSIONS
The above histologic parameters can be helpful in predicting ALK rearranged pulmonary adenocarcinoma, leading to rapid FISH analysis and timely treatment.
Keyword
MeSH Terms
Figure
Cited by 1 articles
-
Non-small Cell Lung Cancer with Concomitant
EGFR ,KRAS , andALK Mutation: Clinicopathologic Features of 12 Cases
Taebum Lee, Boram Lee, Yoon-La Choi, Joungho Han, Myung-Ju Ahn, Sang-Won Um
J Pathol Transl Med. 2016;50(3):197-203. doi: 10.4132/jptm.2016.03.09.
Reference
-
1. Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007; 448:561–6.2. Sun JM, Lira M, Pandya K, et al. Clinical characteristics associated with ALK rearrangements in never-smokers with pulmonary adenocarcinoma. Lung Cancer. 2014; 83:259–64.3. O’Bryant CL, Wenger SD, Kim M, Thompson LA. Crizotinib: a new treatment option for ALK-positive non-small cell lung cancer. Ann Pharmacother. 2013; 47:189–97.
Article4. Paik JH, Choe G, Kim H, et al. Screening of anaplastic lymphoma kinase rearrangement by immunohistochemistry in non-small cell lung cancer: correlation with fluorescence in situ hybridization. J Thorac Oncol. 2011; 6:466–72.5. Yi ES, Boland JM, Maleszewski JJ, et al. Correlation of IHC and FISH for ALK gene rearrangement in non-small cell lung carcinoma: IHC score algorithm for FISH. J Thorac Oncol. 2011; 6:459–65.6. Yoshida A, Tsuta K, Nakamura H, et al. Comprehensive histologic analysis of ALK-rearranged lung carcinomas. Am J Surg Pathol. 2011; 35:1226–34.7. Jokoji R, Yamasaki T, Minami S, et al. Combination of morphological feature analysis and immunohistochemistry is useful for screening of EML4-ALK-positive lung adenocarcinoma. J Clin Pathol. 2010; 63:1066–70.8. Inamura K, Takeuchi K, Togashi Y, et al. EML4-ALK fusion is linked to histological characteristics in a subset of lung cancers. J Thorac Oncol. 2008; 3:13–7.9. Inamura K, Takeuchi K, Togashi Y, et al. EML4-ALK lung cancers are characterized by rare other mutations, a TTF-1 cell lineage, an acinar histology, and young onset. Mod Pathol. 2009; 22:508–15.10. Rodig SJ, Mino-Kenudson M, Dacic S, et al. Unique clinicopathologic features characterize ALK-rearranged lung adenocarcinoma in the western population. Clin Cancer Res. 2009; 15:5216–23.11. Kim H, Jang SJ, Chung DH, et al. A comprehensive comparative analysis of the histomorphological features of ALK-rearranged lung adenocarcinoma based on driver oncogene mutations: frequent expression of epithelial-mesenchymal transition markers than other genotype. PLoS One. 2013; 8:e76999.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Anaplastic lymphoma kinase (ALK)-expressing Lung Adenocarcinoma with Combined Neuroendocrine Component or Neuroendocrine Transformation: Implications for Neuroendocrine Transformation and Response to ALK-tyrosine Kinase Inhibitors
- Histologic Features of ALK-Expressing Adenocarciomas of the Lung
- A Case of Multiple Cranial Neuropathies Caused by Anaplastic Lymphoma Kinase-Negative Anaplastic Large Cell Lymphoma
- CD30-Positive Anaplastic Lymphoma Kinase-Negative Systemic Anaplastic Large-Cell Lymphoma in a 9-Year-Old Boy
- Cytologic Features of ALK-Positive Pulmonary Adenocarcinoma