Ann Lab Med.
2014 Mar;34(2):139-144. 10.3343/alm.2014.34.2.139.
Antimicrobial Resistance Patterns and Integron Carriage of Escherichia coli Isolates Causing Community-Acquired Infections in Turkey
- Affiliations
-
- 1Department of Medical Microbiology, Faculty of Medicine, Recep Tayyip Erdogan University, Rize, Turkey. draysegulcicek@yahoo.com
- 2Department of Biology, Faculty of Arts & Sciences, Artvin Coruh University, Artvin, Turkey.
- 3Department of Biology, Faculty of Arts & Sciences, Recep Tayyip Erdogan University, Rize, Turkey.
- KMID: 1791907
- DOI: http://doi.org/10.3343/alm.2014.34.2.139
Abstract
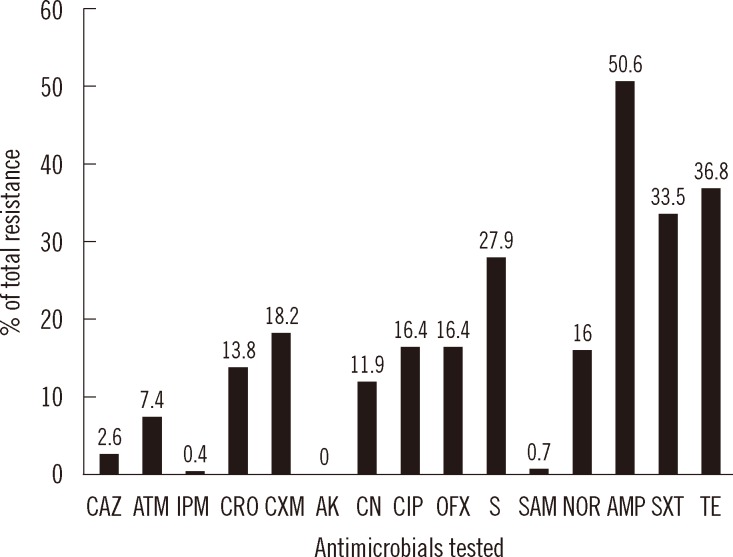
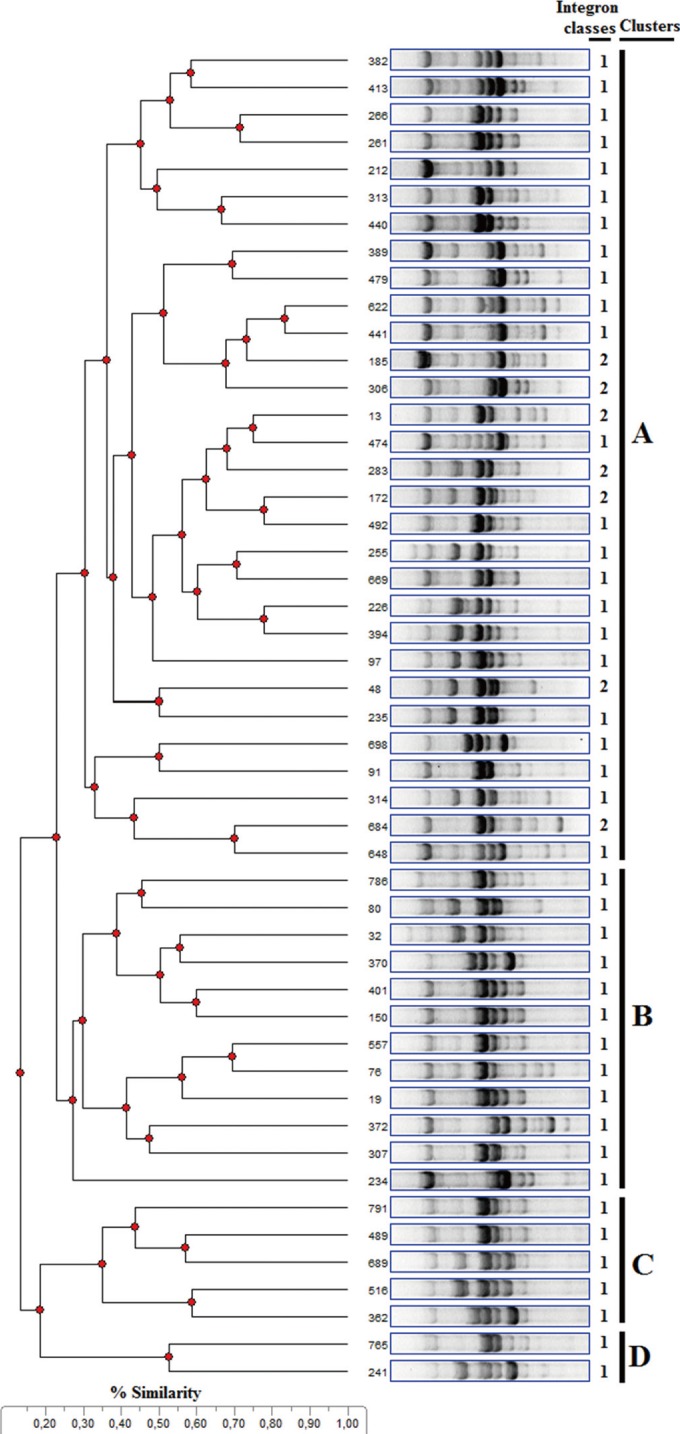
- We aimed to observe antimicrobial resistance patterns and integron carriage of Escherichia coli isolates causing community-acquired infections. Two hundred sixty-eight E. coli strains were obtained from outpatients with various infections at different polyclinics at the 82nd Year of State Hospital in Rize, Turkey. Susceptibility to antimicrobials was tested using a disk diffusion method. The presence of integrons was examined using PCR with specific primers. Positive PCR results were confirmed by sequencing. A broth mating method was used for conjugation assays. Extragenic palindromic-PCR was performed using the oligonucleotide primer BOXA1R. Resistance frequency for ampicillin, trimethoprim/sulfamethoxazole, and tetracycline was determined as 50.6%, 33.5%, and 36.8% respectively. No strains were resistant to amikacin. Seventy isolates were positive for the intI1 gene, of which 49 carried gene cassettes. Eleven isolates were positive for the intI2 gene, eight of which carried gene cassettes. Seven gene cassettes (dfrA1, dfrA5, dfrA7, dfrA17, aadA1, aadA5, and sat2) were predominantly harbored in integrons. We detected conjugative plasmids harboring integrons in two E. coli strains. Four strain clusters were yielded by BOX-PCR fingerprints showing that they were clonally related. No apparent relationship occurred among class 1 and 2 integron-carrying strains. We conclude that integrons are widespread in genetically variable E. coli strains and will continue to mediate dissemination of resistance genes in the community.
MeSH Terms
-
Anti-Bacterial Agents/*pharmacology
Community-Acquired Infections/*microbiology
Disk Diffusion Antimicrobial Tests
Drug Resistance, Bacterial
Escherichia coli/*drug effects/isolation & purification
Escherichia coli Proteins/*genetics
Humans
Integrases/genetics
Polymerase Chain Reaction
Turkey
Anti-Bacterial Agents
Escherichia coli Proteins
Integrases
Figure
Reference
-
1. Jones RN, Kugler KC, Pfaller MA, Winokur PL. Characteristics of pathogens causing urinary tract infections in hospitals in North America: results from the SENTRY Antimicrobial Surveillance Program, 1997. Diagn Microbiol Infect Dis. 1999; 35:55–63. PMID: 10529882.
Article2. Rice LB. The clinical consequences of antimicrobial resistance. Curr Opin Microbiol. 2009; 12:476–481. PMID: 19716760.
Article3. Cocchi S, Grasselli E, Gutacker M, Benagli C, Convert M, Piffaretti JC. Distribution and characterization of integrons in Escherichia coli strains of animal and human origin. FEMS Immunol Med Microbiol. 2007; 50:126–132. PMID: 17456180.4. Sandalli C, Buruk CK, Sancaktar M, Ozgumus OB. Prevalence of integrons and a new dfrA17 variant in Gram-negative bacilli that cause community-acquired infections. Microbiol Immunol. 2010; 54:164–169. PMID: 20236427.5. National Committee for Clinical Laboratory Standards. Tenth Informational supplement, M100-S10 (M2). Performance standards for antimicrobial susceptibility testing. Wayne, PA: National Committee for Clinical Laboratory Standards;2000.6. Lévesque C, Piché L, Larose C, Roy PH. PCR Mapping of integrons reveals several novel combinations of resistance genes. Antimicrob Agents Chemother. 1995; 39:185–191. PMID: 7695304.
Article7. White PA, McIver CJ, Rawlinson WD. Integrons and gene cassettes in the enterobacteriaceae. Antimicrob Agents Chemother. 2001; 45:2658–2661. PMID: 11502548.9. Rice LB, Willey SH, Papanicolaou GA, Medeiros AA, Eliopoulos GM, Moellering RC Jr, et al. Outbreak of ceftazidime resistance caused by extended-spectrum β-lactamases at a Massachusetts chronic-care facility. Antimicrob Agents Chemother. 1990; 34:2193–2199. PMID: 2073110.10. Versalovic J, Schneider M, de Bruijn FJ, Lupski JR. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol Cell Biol. 1994; 5:25–40.11. Seurinck S, Verstraete W, Siciliano SD. Use of 16S-23S rRNA intergenic spacer region PCR and repetitive extragenic palindromic PCR analyses of Escherichia coli isolates to identify nonpoint fecal sources. Appl Environ Microbiol. 2003; 69:4942–4950. PMID: 12902290.12. El-Najjar NG, Farah MJ, Hashwa FA, Tokajian ST. Antibiotic resistance patterns and sequencing of class I integron from uropathogenic Escherichia coli in Lebanon. Lett Appl Microbiol. 2010; 51:456–461. PMID: 20840552.13. Sawma-Aouad G, Hashwa F, Tokajian S. Antimicrobial resistance in relation to virulence determinants and phylogenetic background among uropathogenic Escherichia coli in Lebanon. J Chemother. 2009; 21:153–158. PMID: 19423467.14. Ozgumus OB, Celik-Sevim E, Alpay-Karaoglu S, Sandalli C, Sevim A. Molecular characterization of antibiotic resistant Escherichia coli strains isolated from tap and spring waters in a coastal region in Turkey. J Microbiol. 2007; 45:379–387. PMID: 17978796.15. Ozgumus OB, Sandalli C, Sevim A, Celik-Sevim E, Sivri N. Class 1 and class 2 integrons and plasmid-mediated antibiotic resistance in coliforms isolated from ten rivers in northern Turkey. J Microbiol. 2009; 47:19–27. PMID: 19229487.
Article16. Solberg OD, Ajiboye RM, Riley LW. Origin of class 1 and 2 integrons and gene cassettes in a population-based sample of uropathogenic Escherichia coli. J Clin Microbiol. 2006; 44:1347–1351. PMID: 16597861.17. Jones LA, McIver CJ, Rawlinson WD, White PA. Polymerase chain reaction screening for integrons can be used to complement resistance surveillance programs. Commun Dis Intell Q Rep. 2003; 27(Suppl):S103–S110. PMID: 12807284.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Antimicrobial Resistance and Integrons Found in Commensal Escherichia coli Isolates from Healthy Humans
- Changes in patterns of antimicrobial susceptibility and class 1 integron carriage among Escherichia coli isolates
- Molecular characteristics of Escherichia coli from bulk tank milk in Korea
- Antimicrobial Resistance in Escherichia coli Isolated from Healthy Volunteers of the Community
- Analysis of Integrons and Antimicrobial Resistances of Multidrug Resistant Escherichia coli Isolated in Korea