J Bacteriol Virol.
2019 Dec;49(4):176-190. 10.4167/jbv.2019.49.4.176.
Analysis of Integrons and Antimicrobial Resistances of Multidrug Resistant Escherichia coli Isolated in Korea
- Affiliations
-
- 1Department of Microbiology, Keimyung University School of Medicine, 1035 Dalgubeol-daero, Dalseo-Gu, Daegu, 42601, Korea. minho@dsmc.or.kr
- KMID: 2468024
- DOI: http://doi.org/10.4167/jbv.2019.49.4.176
Abstract
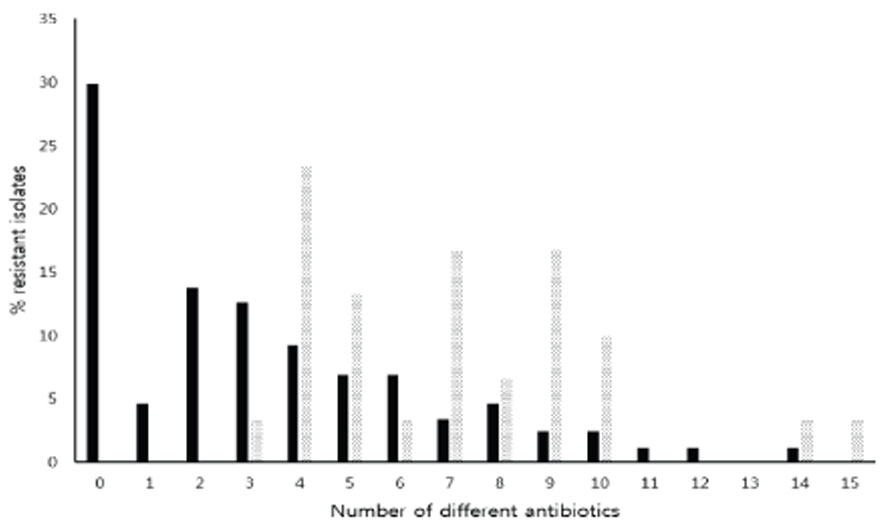
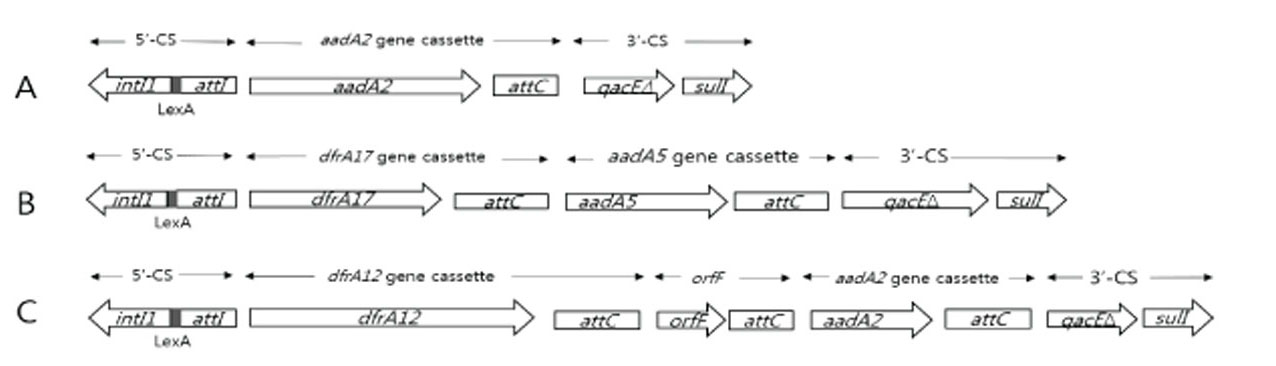
- Antibacterial drugs are one of the most important therapeutic agents of bacterial infections but multidrug resistant Escherichia coli (MDREC) is an increasing problem worldwide. Major resistance mechanism of MDREC is horizontal gene transfer of R plasmids harboring integrons, which the integron integrase (IntI) catalyzes gene cassette insertion and excision through site specific recombination. In this study, resistance profiles of integron harboring E. coli isolated in Korea and the genetic environments of integron gene cassettes were analyzed by PCR and direct sequencing to clarify the mechanisms of spread of integron harboring E. coli. Resistance rates of integron harboring E. coli, including β-lactams, aminoglycosides, and fluoroquinolones and MDR frequencies were significantly higher than that of E. coli without integron (p < 0.01). Majority (80%) of integron harboring E. coli showed resistance transfer by conjugation. Most (80%) of E. coli had dfrA17-aadA5 cassette array and PcH1 hybrid promoter; 16.7% of E. coli had dfrA12-orfF-aadA2 cassette array and PcW promoter. The higher prevalence of weak Pc variants among most (96.7%) of integron harboring MDREC suggests that a flexible cassette array is more important than enhanced expression. All the integrons had LexA binding motif suggests that SOS responses control the expression of these integrons. In conclusion, the genetic bases of integrons were diverse, and the spread and the expression of prevalent gene cassette arrays may be deeply related with strengths of Pc promoters in integrons. These informations will provide important knowledge to control the increase of integron harboring MDREC.
Keyword
MeSH Terms
Figure
Reference
-
1. Rogers BA, Sidjabat HE, Paterson DL. Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J Antimicrob Chemother. 2011; 66:1–14.
Article2. Naseer U, Sundsfjord A. The CTX-M conundrum: dissemination of plasmids and Escherichia coli clones. Microb Drug Resist. 2011; 17:83–97.
Article3. Yang YY, Suh MH. Diversity of genetic environment of blaCTX-M genes and antimicrobial susceptibility in extendedspectrum β-lactamase producing Escherichia coli isolated in Korea. J Bacteriol Virol. 2019; 49:95–114.
Article4. Carattoli A. Plasmids and the spread of resistance. Int J Med Microbiol. 2013; 303:298–304.
Article5. Sunde M, Simonsen GS, Slettemeås JS, Böckerman I, Norström M. Integron, plasmid and host strain characteristics of Escherichia coli from humans and food included in the Norwegian antimicrobial resistance monitoring programs. PLoS One. 2015; 10:e0128797.6. Escudero JA, Loot C, Nivina A, Mazel D. The Integron: Adaptation on demand. In : Craig NL, editor. Mobile DNA III. 3rd ed. Washington, DC: ASM Press;2015. p. 139–161.7. Hickman AB, Dyda F. Mechanisms of DNA transposition. In : Craig NL, editor. Mobile DNA III. 3rd ed. Washington, DC: ASM Press;2015. p. 531–553.8. Partridge SR, Tsafnat G, Coiera E, Iredell JR. Gene cassettes and cassette arrays in mobile resistance integrons. FEMS Microbiol Rev. 2009; 33:757–784.
Article9. Gillings MR. Integrons: past, present, and future. Microbiol Mol Biol Rev. 2014; 78:257–277.
Article10. Jové T, Da Re S, Denis F, Mazel D, Ploy MC. Inverse correlation between promoter strength and excision activity in class 1 integrons. PLoS genet. 2010; 6:e1000793.
Article11. Vinué L, Jové T, Torres C, Ploy MC. Diversity of class 1 integron gene cassette Pc promoter variants in clinical Escherichia coli strains and description of a new P2 promoter variant. Int J Antimicrob Agents. 2011; 38:526–529.
Article12. Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing. 24th ed. Wayne: CLSI Press;2014.13. Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012; 18:268–281.
Article14. Kado CI, Liu ST. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981; 145:1365–1373.
Article15. Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005; 63:219–228.
Article16. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003; 31:3406–3415.
Article17. El Salabi A, Walsh TR, Chouchani C. Extended spectrum beta-lactamases, carbapenemases and mobile genetic elements responsible for antibiotics resistance in Gram-negative bacteria. Crit Rev Microbiol. 2013; 39:113–122.
Article18. Pitout JD, Laupland KB. Extended-spectrum β-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect Dis. 2008; 8:159–166.
Article19. Ruiz del Castillo B, Vinué L, Román EJ, Guerra B, Carattoli A, Torres C, et al. Molecular characterization of multiresistant Escherichia coli producing or not extended-spectrum β-lactamases. BMC Microbiol. 2013; 13:84.
Article20. Li B, Zhao ZC, Wang MH, Huang XH, Pan YH, Cao YP. Antimicrobial resistance and integrons of commensal Escherichia coli strains from healthy humans in China. J Chemother. 2014; 26:190–192.
Article21. Kang HY, Jeong YS, Oh JY, Tae SH, Choi CH, Moon DC, et al. Characterization of antimicrobial resistance and class 1 integrons found in Escherichia coli isolates from humans and animals in Korea. J Antimicrob Chemother. 2005; 55:639–644.
Article22. Shin HW, Lim J, Kim S, Kim J, Kwon GC, Koo SH. Characterization of trimethoprim-sulfamethoxazole resistance genes and their relatedness to class 1 integron and insertion sequence common region in gram-negative bacilli. J Microbiol Biotechnol. 2015; 25:137–142.
Article23. Martines-Freijo P, Fluit AC, Schmitz FJ, Grek VS, Verhoef J, Jones ME. Class I integrons in gram-negative isolates from different European hospitals and association with decreased susceptibility to multiple antibiotic compounds. J Antimicrob Chemother. 1998; 42:689–696.
Article24. Gillings M, Boucher Y, Labbate M, Holmes A, Krishnan S, Holley M, et al. The evolution of class 1 integrons and the rise of antibiotic resistance. J Bacteriol. 2008; 190:5095–5100.
Article25. Yu HS, Lee JC, Kang HY, Jeong YS, Lee EY, Choi CH, et al. Prevalence of dfr genes associated with integrons and dissemination of dfrA17 among urinary isolates of Escherichia coli in Korea. J Antimicrob Chemother. 2004; 53:445–450.
Article26. Chen T, Feng Y, Yuan JL, Qi Y, Cao YX, Wu Y. Class 1 integrons contributes to antibiotic resistance among clinical isolates of Escherichia coli producing extended-spectrum beta-lactamases. Indian J Med Microbiol. 2013; 31:385–389.
Article27. Seo KW, Lee YJ. Prevalence and characterization of β-lactamase genes and class 1 integrons in multidrug-resistant Escherichia coli isolates from chicken meat in Korea. Microb Drug Resist. 2018.28. Gu B, Pan S, Wang T, Zhao W, Mei Y, Huang P, et al. Novel cassette arrays of integrons in clinical strains of Enterobacteriaceae in China. Int J Antimibrob Agents. 2008; 32:529–533.
Article29. Levy-Hara G, Amábile-Cuevas CF, Gould I, Hutchinson J, Abbo L, Saxynger L, et al. “Ten commandments” for the appropriate use of antibiotics by the practicing physician in an outpatient setting. Front Microbiol. 2011; 2:230.30. Bush K. Alarming β-lactamase-mediated resistance in multidrug-resistant Enterobacteriaceae. Curr Opin Microbiol. 2010; 13:558–564.
Article31. Cantón R, Morosini M. Emergence and spread of antibiotic resistance following exposure to antibiotics. FEMS Microbiol Rev. 2011; 35:977–991.
Article32. Kuo HY, Chang KC, Kuo JW, Yueh HW, Liou ML. Imipenem: a potent inducer of multidrug resistance in Acinetobacter baumannii. Int J Antimicrob Agents. 2012; 39:33–38.
Article33. Villa L, García-Fernández A, Fortini D, Carattoli A. Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J Antimicrob Chemother. 2010; 65:2518–2529.
Article34. Bouvier M, Ducos-Galand M, Loot C, Bikard D, Mazel D. Structural features of single-stranded integron cassette attC sites and their role in strand selection. PLoS genet. 2009; 5:e1000632.
Article35. Cambray G, Sanchez-Alberola N, Campoy S, Guerin E, Da Re S, González-Zorn B, et al. Prevalence of SOS-mediated control of integron integrase expression as an adaptive trait of chromosomal and mobile integrons. Mob DNA. 2011; 2:6.
Article36. Lu TK, Collins JJ. Engineered bacteriophage targeting gene networks as adjuvants for antibiotic therapy. Proc Natl Acad Sci U S A. 2009; 106:4629–4634.
Article37. López E, Blázquez J. Effect of subinhibitory concentrations of antibiotics on intrachromosomal homologous recombination in Escherichia coli. Antimicrob Agents Chemother. 2009; 53:3411–3415.
Article38. Hocquet D, Bertrand X. Metronidazole increases the emergence of ciprofloxacin- and amikacin-resistant Pseudomonas aeruginosa by inducing the SOS response. J Antimicrob Chemother. 2014; 69:852–854.
Article39. Baharoglu Z, Bikard D, Mazel D. Conjugative DNA transfer induces the bacterial SOS response and promotes antibiotic resistance development through integron activation. PLoS Genet. 2010; 6:e1001165.
Article40. Strugeon E, Tilloy V, Ploy MC, Da Re S. The stringent response promotes antibiotic resistance dissemination by regulating integron integrase expression in biofilms. MBio. 2016; 7.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Antimicrobial Resistance and Integrons Found in Commensal Escherichia coli Isolates from Healthy Humans
- High Prevalence of Class 1 to 3 Integrons Among Multidrug-Resistant Diarrheagenic
Escherichia coli in Southwest of Iran - Changes in patterns of antimicrobial susceptibility and class 1 integron carriage among Escherichia coli isolates
- Comparison of Antimicrobial Resistance in
Escherichia coli Strains Isolated From Healthy Poultry and Swine Farm Workers Using Antibiotics in Korea - Antimicrobial Resistance Patterns and Integron Carriage of Escherichia coli Isolates Causing Community-Acquired Infections in Turkey