J Korean Med Sci.
2007 Sep;22(Suppl):S32-S37. 10.3346/jkms.2007.22.S.S32.
Hypermethylation of p16(INK4a) in Korean Non-small Cell Lung Cancer Patients
- Affiliations
-
- 1Department of Preventive Medicine, College of Medicine, Dong-A University, Busan, Korea. yshong@dau.ac.kr
- 2Department of Pathology, College of Medicine, Dong-A University, Busan, Korea.
- 3Medical Research Center for Cancer Molecular Therapy, College of Medicine, Dong-A University, Busan, Korea.
- KMID: 1785786
- DOI: http://doi.org/10.3346/jkms.2007.22.S.S32
Abstract
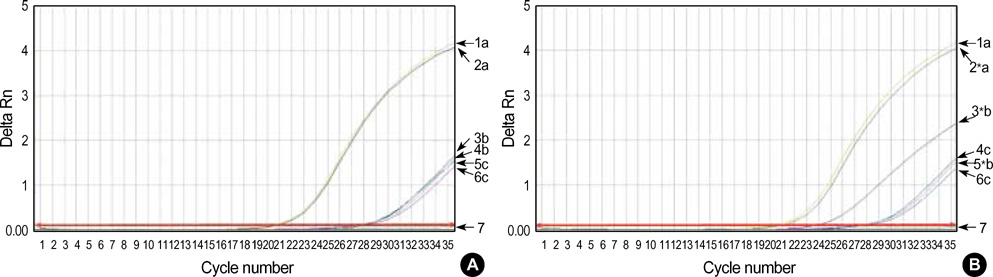
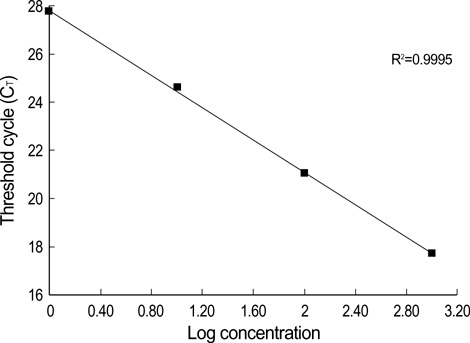
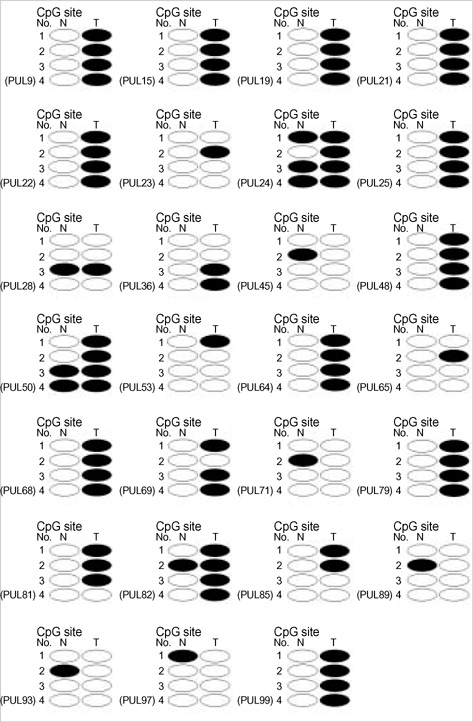
- Promoter hypermethylation of the p16(INK4a) gene was investigated in 81 sets of samples of tumor tissue and adjacent normal tissue from Korean patients with primary lung cancer, using the modified real-time polymerase chain reaction (PCR)/ SYBR Green detection method. The results showed hypermethylation of p16(INK4a) in 27.2% of tumor tissues, and in 11.1% of adjacent normal tissue. No significant association was found between the overall aberrant methylation in tumor and corresponding normal specimens (r=0.137, p=0.219). In 22 cases with p16(INK4a) hypermethylation in tumor tissues, only 4 (18.1%) cases were found to have a hypermethylated normal tissue specimen. The findings of this study show that smoking can influence the methylation level of the promoter region of p16(INK4a), and that this occurs in tumor tissues more frequently than in normal tissues. Other clinicopathological characteristics, including age, sex, tumor stage, and histologic type were not found to be correlated with p16(INK4a) methylation.
Keyword
MeSH Terms
-
Adult
Aged
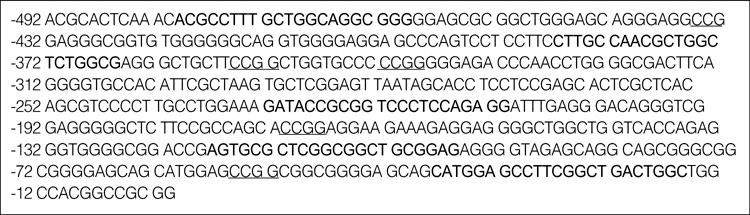
Base Sequence
Carcinoma, Non-Small-Cell Lung/*genetics
*DNA Methylation
DNA Primers/genetics
DNA, Neoplasm/chemistry/genetics
Female
*Genes, p16
Humans
Korea
Lung Neoplasms/*genetics
Male
Middle Aged
Molecular Sequence Data
Polymerase Chain Reaction
Promoter Regions, Genetic
Smoking/adverse effects/genetics
Figure
Reference
-
1. Monk M. Epigenetic programming of differential gene expression in development and evolution. Dev Genet. 1995. 17:188–197.
Article2. Turker MS, Bestor TH. Formation of methylation patterns in the mammalian genome. Mutat Res. 1997. 386:119–130.
Article3. Paulsen M, Ferguson-Smith AC. DNA methylation in genomic imprinting, development and disease. J Pathol. 2001. 195:97–110.
Article4. Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res. 1998. 72:141–196.5. Gray SG, Eriksson T, Ekstrom TJ. Methylation, gene expression and the chromatin connection in cancer (review). Int J Mol Med. 1999. 4:333–350.
Article6. Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001. 61:3225–3229.7. Harden SV, Tokumaru Y, Westra WH, Goodman S, Ahrendt SA, Yang SC, Sidransky D. Gene promoter hypermethylation in tumors and lymph nodes of stage I lung cancer patients. Clin Cancer Res. 2003. 9:1370–1375.8. Liu Y, An Q, Li L, Zhang D, Huang J, Feng X, Cheng S, Gao Y. Hypermethylation of p16INK4a in Chinese lung cancer patients: biological and clinical implications. Carcinogenesis. 2003. 24:1897–1901.
Article9. Ulivi P, Zoli W, Calistri D, Fabbri F, Tesei A, Rosetti M, Mengozzi M, Amadori D. p16INK4A and CDH13 hypermethylation in tumor and serum of non-small cell lung cancer patients. J Cell Physiol. 2006. 206:611–615.10. Chu DC, Chuang CK, Fu JB, Huang HS, Tseng CP, Sun CF. The use of real-time quantitative polymerase chain reaction to detect hypermethylation of the CpG islands in the promoter region flanking the GSTP1 gene to diagnose prostate carcinoma. J Urol. 2002. 167:1854–1858.
Article11. Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1997. 37:646–650.12. Nakayama H, Hibi K, Takase T, Yamazaki T, Kasai Y, Ito K, Akiyama S, Nakao A. Molecular detection of p16 promoter methylation in the serum of recurrent colorectal cancer patients. Int J Cancer. 2003. 105:491–493.
Article13. Reed AL, Califano J, Cairns P, Westra WH, Jones RM, Koch W, Ahrendt S, Eby Y, Sewell D, Nawroz H, Bartek J, Sidransky D. High frequency of p16 (CDKN2/MTS-1/INK4A) inactivation in head and neck squamous cell carcinoma. Cancer Res. 1994. 56:3630–3633.14. Stroun M, Anker P, Maurice P, Lyautey J, Lederrey C, Beljanski M. Neoplastic characteristics of the DNA found in the plasma of cancer patients. Oncology. 1989. 46:318–322.
Article15. Wong TS, Man MW, Lam AK, Wei WI, Kwong YL, Yuen AP. The study of p16 and p15 gene methylation in head and neck squamous cell carcinoma and their quantitative evaluation in plasma by real-time PCR. Eur J Cancer. 2003. 39:1881–1887.
Article16. Singer-Sam J, LeBon JM, Tanguay RL, Riggs AD. A quantitative HpaII-PCR assay to measure methylation of DNA from a small number of cells. Nucleic Acids Res. 1990. 18:687.17. Eads CA, Danenberg KD, Kawakami K, Saltz LB, Blake C, Shibata D, Danenberg PV, Laird PW. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000. 28:E32.
Article18. Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci USA. 1992. 89:1827–1831.
Article19. Kim DH, Nelson HH, Wiencke JK, Zheng S, Christiani DC, Wain JC, Mark EJ, Kelsey KT. p16(INK4a) and histology-specific methylation of CpG islands by exposure to tobacco smoke in non-small cell lung cancer. Cancer Res. 2001. 61:3419–3424.20. Jarmalaite S, Kannio A, Anttila S, Lazutka JR, Husgafvel-Pursiainen K. Aberrant p16 promoter methylation in smokers and former smokers with nonsmall cell lung cancer. Int J Cancer. 2003. 106:913–918.21. Kim DH, Kim JS, Ji YI, Shim YM, Kim H, Han J, Park J. Hypermethylation of RASSF1A promoter is associated with the age at starting smoking and a poor prognosis in primary non-small cell lung cancer. Cancer Res. 2003. 63:3743–3746.22. Rom WN, Hay JG, Lee TC, Jiang Y, Tchou-Wong KM. Molecular and genetic aspects of lung cancer. Am J Respir Crit Care Med. 2000. 161:1355–1367.
Article23. Belinsky SA, Nikula KJ, Baylin SB, Issa JP. Increased cytosine DNA-methyltransferase activity is target-cell-specific and an early event in lung cancer. Proc Natl Acad Sci USA. 1996. 93:4045–4050.
Article24. Vertino PM, Yen RW, Gao J, Baylin SB. De novo methylation of CpG island sequences in human fibroblasts overexpressing DNA (cytosine-5-)-methyltransferase. Mol Cell Biol. 1996. 16:4555–4565.
Article25. An Q, Liu Y, Gao Y, Huang J, Fong X, Li L, Zhang D, Cheng S. Detection of p16 hypermethylation in circulating plasma DNA of non-small cell lung cancer patients. Cancer Lett. 2002. 188:109–114.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- p16(INK4a) Promoter Hypermethylation in Sputum, Blood, and Tissue from Non-Small Cell Lung Cancer and Pulmonary Inflammation
- p16(INK4A) promoter hypermethylation and expression of p16(INK4A), cyclin D1, and Rb in papillary thyroid carcinoma
- The p16INK4A Expression in Stomach Cancer , Colon Cancer and Hepatoma Cell Lines
- Inactivation patterns of p16/INK4A in oral squamous cell carcinomas
- The effects of transferring tumor suppressor gene p16INK4A to p16INK4A-deleted cancer cells