Cancer Res Treat.
2023 Jul;55(3):778-803. 10.4143/crt.2022.1042.
MLL4 Regulates the Progression of Non–Small-Cell Lung Cancer by Regulating the PI3K/AKT/SOX2 Axis
- Affiliations
-
- 1Key Laboratory of Imaging Diagnosis and Minimally Invasive Intervention Research, Institute of Imaging Diagnosis and Minimally Invasive Intervention Research, The Fifth Affiliated Hospital of Wenzhou Medical University, Lishui, China
- 2Department of Radiology, Clinical College of The Affiliated Central Hospital, Lishui University, Lishui, China
- 3Department of Radiology, Lishui Hospital of Zhejiang University, Lishui, China
- KMID: 2544161
- DOI: http://doi.org/10.4143/crt.2022.1042
Abstract
- Purpose
Mixed-lineage leukemia protein 4 (MLL4/KMT2D) is a histone methyltransferase, and its mutation has been reported to be associated with a poor prognosis in many cancers, including lung cancer. We investigated the function of MLL4 in lung carcinogenesis.
Materials and Methods
RNA sequencing (RNA-seq) in A549 cells transfected with control siRNA or MLL4 siRNA was performed. Also, we used EdU incorporation assay, colony formation assays, growth curve analysis, transwell invasion assays, immunohistochemical staining, and in vivo bioluminescence assay to investigate the function of MLL4 in lung carcinogenesis.
Results
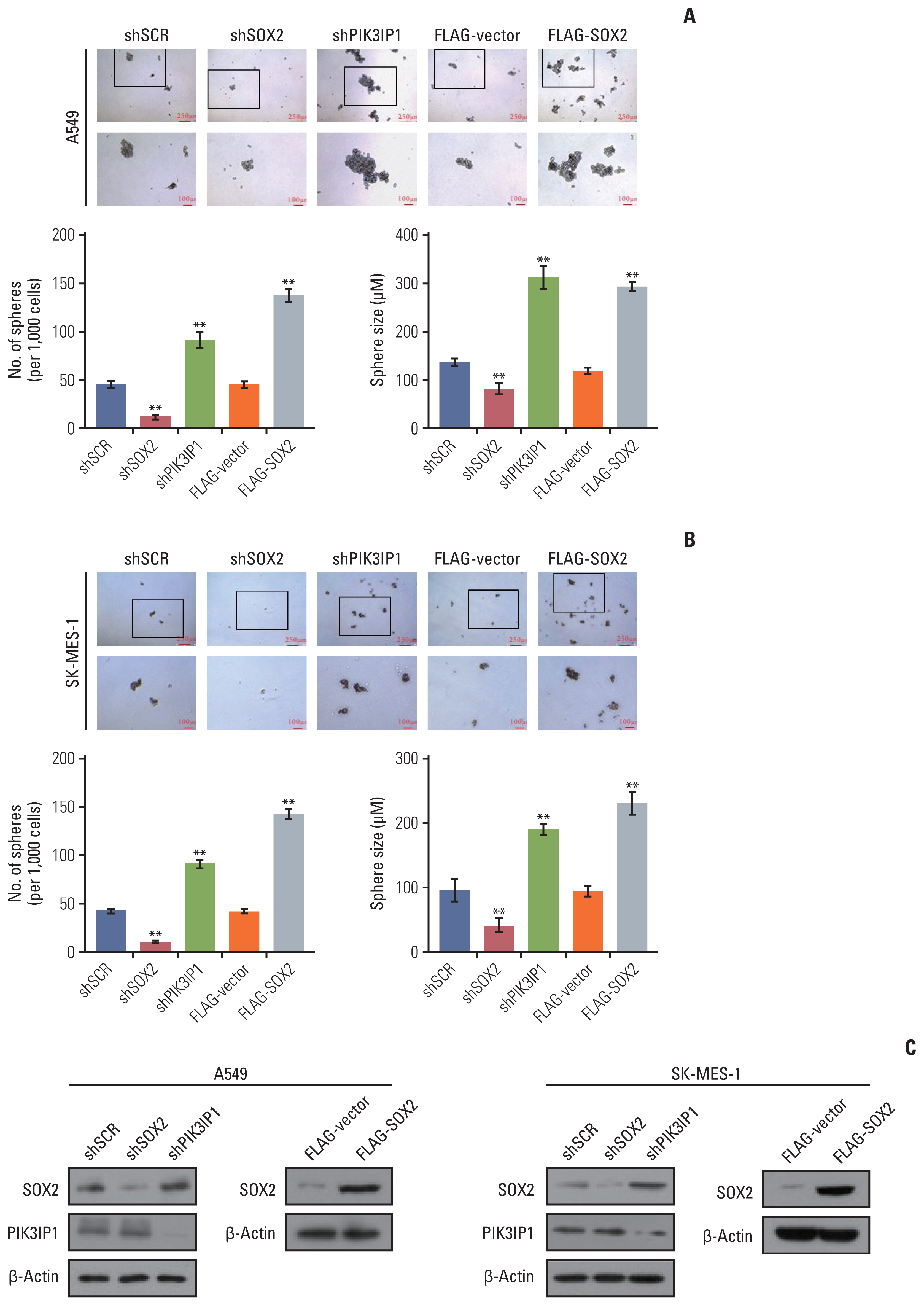
We found that MLL4 expression was downregulated in non–small cell lung cancer (NSCLC) tissues compared to adjacent normal tissues and tended to decrease with disease stage progression. We analyzed the transcriptomes in control and MLL4- deficient cells using high-throughput RNA deep sequencing (RNA-seq) and identified a cohort of target genes, such as SOX2, ATF1, FOXP4, PIK3IP1, SIRT4, TENT5B, and LFNG, some of which are related to proliferation and metastasis. Our results showed that low expression of MLL4 promotes NSCLC cell proliferation and metastasis and is required for the maintenance of NSCLC stem cell properties.
Conclusion
Our findings identify an important role of MLL4 in lung carcinogenesis through transcriptional regulation of PIK3IP1, affecting the PI3K/AKT/SOX2 axis, and suggest that MLL4 could be a potential prognostic indicator and target for NSCLC therapy.
Keyword
Figure
Reference
-
References
1. Maiuthed A, Chantarawong W, Chanvorachote P. Lung cancer stem cells and cancer stem cell-targeting natural compounds. Anticancer Res. 2018; 38:3797–809.
Article2. Yang S, Zhang Z, Wang Q. Emerging therapies for small cell lung cancer. J Hematol Oncol. 2019; 12:47.
Article3. Dong S, Li W, Wang L, Hu J, Song Y, Zhang B, et al. Histone-related genes are hypermethylated in lung cancer and hypermethylated HIST1H4F could serve as a pan-cancer biomarker. Cancer Res. 2019; 79:6101–12.4. Fagan RJ, Dingwall AK. COMPASS ascending: emerging clues regarding the roles of MLL3/KMT2C and MLL2/KMT-2D proteins in cancer. Cancer Lett. 2019; 458:56–65.
Article5. Gonda TJ, Ramsay RG. Directly targeting transcriptional dysregulation in cancer. Nat Rev Cancer. 2015; 15:686–94.6. Ford DJ, Dingwall AK. The cancer COMPASS: navigating the functions of MLL complexes in cancer. Cancer Genet. 2015; 208:178–91.7. Ardeshir-Larijani F, Bhateja P, Lipka MB, Sharma N, Fu P, Dowlati A. KMT2D mutation is associated with poor prognosis in non-small-cell lung cancer. Clin Lung Cancer. 2018; 19:e489–501.
Article8. Lin-Shiao E, Lan Y, Coradin M, Anderson A, Donahue G, Simpson CL, et al. KMT2D regulates p63 target enhancers to coordinate epithelial homeostasis. Genes Dev. 2018; 32:181–93.9. Froimchuk E, Jang Y, Ge K. Histone H3 lysine 4 methyltransferase KMT2D. Gene. 2017; 627:337–42.10. Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007; 25:15–30.11. Dawkins JB, Wang J, Maniati E, Heward JA, Koniali L, Kocher HM, et al. Reduced expression of histone methyltransferases KMT2C and KMT2D correlates with improved outcome in pancreatic ductal adenocarcinoma. Cancer Res. 2016; 76:4861–71.12. Lv S, Ji L, Chen B, Liu S, Lei C, Liu X, et al. Histone methyltransferase KMT2D sustains prostate carcinogenesis and metastasis via epigenetically activating LIFR and KLF4. Oncogene. 2018; 37:1354–68.
Article13. Hillman RT, Celestino J, Terranova C, Beird HC, Gumbs C, Little L, et al. KMT2D/MLL2 inactivation is associated with recurrence in adult-type granulosa cell tumors of the ovary. Nat Commun. 2018; 9:2496.
Article14. Sun P, Wu T, Sun X, Cui Z, Zhang H, Xia Q, et al. KMT2D inhibits the growth and metastasis of bladder cancer cells by maintaining the tumor suppressor genes. Biomed Pharmacother. 2019; 115:108924.15. Ortega-Molina A, Boss IW, Canela A, Pan H, Jiang Y, Zhao C, et al. The histone lysine methyltransferase KMT2D sustains a gene expression program that represses B cell lymphoma development. Nat Med. 2015; 21:1199–208.
Article16. Xiong W, Deng Z, Tang Y, Deng Z, Li M. Downregulation of KMT2D suppresses proliferation and induces apoptosis of gastric cancer. Biochem Biophys Res Commun. 2018; 504:129–36.
Article17. Alam H, Tang M, Maitituoheti M, Dhar SS, Kumar M, Han CY, et al. KMT2D deficiency impairs super-enhancers to confer a glycolytic vulnerability in lung cancer. Cancer Cell. 2020; 37:599–617.
Article18. Zeng Y, Qiu R, Yang Y, Gao T, Zheng Y, Huang W, et al. Regulation of EZH2 by SMYD2-mediated lysine methylation is implicated in tumorigenesis. Cell Rep. 2019; 29:1482–98.
Article19. Yang Y, Qiu R, Zhao S, Shen L, Tang B, Weng Q, et al. SMYD3 associates with the NuRD (MTA1/2) complex to regulate transcription and promote proliferation and invasiveness in hepatocellular carcinoma cells. BMC Biol. 2022; 20:294.
Article20. Sikorska M, Sandhu JK, Deb-Rinker P, Jezierski A, Leblanc J, Charlebois C, et al. Epigenetic modifications of SOX2 enhancers, SRR1 and SRR2, correlate with in vitro neural differentiation. J Neurosci Res. 2008; 86:1680–93.21. Ediriweera MK, Tennekoon KH, Samarakoon SR. Role of the PI3K/AKT/mTOR signaling pathway in ovarian cancer: biological and therapeutic significance. Semin Cancer Biol. 2019; 59:147–60.
Article22. Tewari D, Patni P, Bishayee A, Sah AN, Bishayee A. Natural products targeting the PI3K-Akt-mTOR signaling pathway in cancer: a novel therapeutic strategy. Semin Cancer Biol. 2022; 80:1–17.
Article23. Yang Q, Jiang W, Hou P. Emerging role of PI3K/AKT in tumor-related epigenetic regulation. Semin Cancer Biol. 2019; 59:112–24.
Article24. Wang Z, Kang L, Zhang H, Huang Y, Fang L, Li M, et al. AKT drives SOX2 overexpression and cancer cell stemness in esophageal cancer by protecting SOX2 from UBR5-mediated degradation. Oncogene. 2019; 38:5250–64.
Article25. Mamun MA, Mannoor K, Cao J, Qadri F, Song X. SOX2 in cancer stemness: tumor malignancy and therapeutic potentials. J Mol Cell Biol. 2020; 12:85–98.
Article26. Karachaliou N, Rosell R, Viteri S. The role of SOX2 in small cell lung cancer, lung adenocarcinoma and squamous cell carcinoma of the lung. Transl Lung Cancer Res. 2013; 2:172–9.27. Singh S, Trevino J, Bora-Singhal N, Coppola D, Haura E, Altiok S, et al. EGFR/Src/Akt signaling modulates Sox2 expression and self-renewal of stem-like side-population cells in non-small cell lung cancer. Mol Cancer. 2012; 11:73.
Article28. Schaefer T, Lengerke C. SOX2 protein biochemistry in stem-ness, reprogramming, and cancer: the PI3K/AKT/SOX2 axis and beyond. Oncogene. 2020; 39:278–92.29. Schaefer T, Ramadoss A, Leu S, Tintignac L, Tostado C, Bink A, et al. Regulation of glioma cell invasion by 3q26 gene products PIK3CA, SOX2 and OPA1. Brain Pathol. 2019; 29:336–50.
Article30. Tang J, Zhong G, Wu J, Chen H, Jia Y. SOX2 recruits KLF4 to regulate nasopharyngeal carcinoma proliferation via PI3K/AKT signaling. Oncogenesis. 2018; 7:61.
Article31. Ormsbee Golden BD, Wuebben EL, Rizzino A. Sox2 expression is regulated by a negative feedback loop in embryonic stem cells that involves AKT signaling and FoxO1. PLoS One. 2013; 8:e76345.
Article32. Egolf S, Zou J, Anderson A, Simpson CL, Aubert Y, Prouty S, et al. MLL4 mediates differentiation and tumor suppression through ferroptosis. Sci Adv. 2021; 7:eabj9141.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Gallic Acid Hindered Lung Cancer Progression by Inducing Cell Cycle Arrest and Apoptosis in A549 Lung Cancer Cells via PI3K/Akt Pathway
- Dual Inhibition of PI3K/Akt/mTOR Pathway and Role of Autophagy in Non-Small Cell Lung Cancer Cells
- Membrane Trafficking of Collecting Duct Water Channel Protein AQP2 Regulated by Akt/AS160
- Up-regulation of P13K/Akt Signaling by 17 beta-estradiol through Activation of Estrogen Receptor-alpha in Breast Cancer Cells
- Long Noncoding RNA PVT1 Promotes Stemness and Temozolomide Resistance through miR-365/ELF4/SOX2 Axis in Glioma