Cancer Res Treat.
2022 Jul;54(3):753-766. 10.4143/crt.2021.905.
The Feasibility of Using Biomarkers Derived from Circulating Tumor DNA Sequencing as Predictive Classifiers in Patients with Small-Cell Lung Cancer
- Affiliations
-
- 1Department of Medical Oncology, National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing, China
- 2Medical Center, Geneplus-Beijing, Beijing, China
- 3Department of Medical Oncology, The People's Hospital of Tangshan city, Tangshan, China
- 4Department of Respiratory and Critical Care Medicine, Beijing Luhe Hospital, Capital Medical University, Beijing, China
- 5Department of General Medicine, Beijing Chest Hospital, Capital Medical University & Beijing Tuberculosis and Thoracic Tumor Research Institute, Beijing, China
- KMID: 2531322
- DOI: http://doi.org/10.4143/crt.2021.905
Abstract
- Purpose
To investigate the feasibility of biomarkers based on dynamic circulating tumor DNA (ctDNA) to classify small cell lung cancer (SCLC) into different subtypes.
Materials and Methods
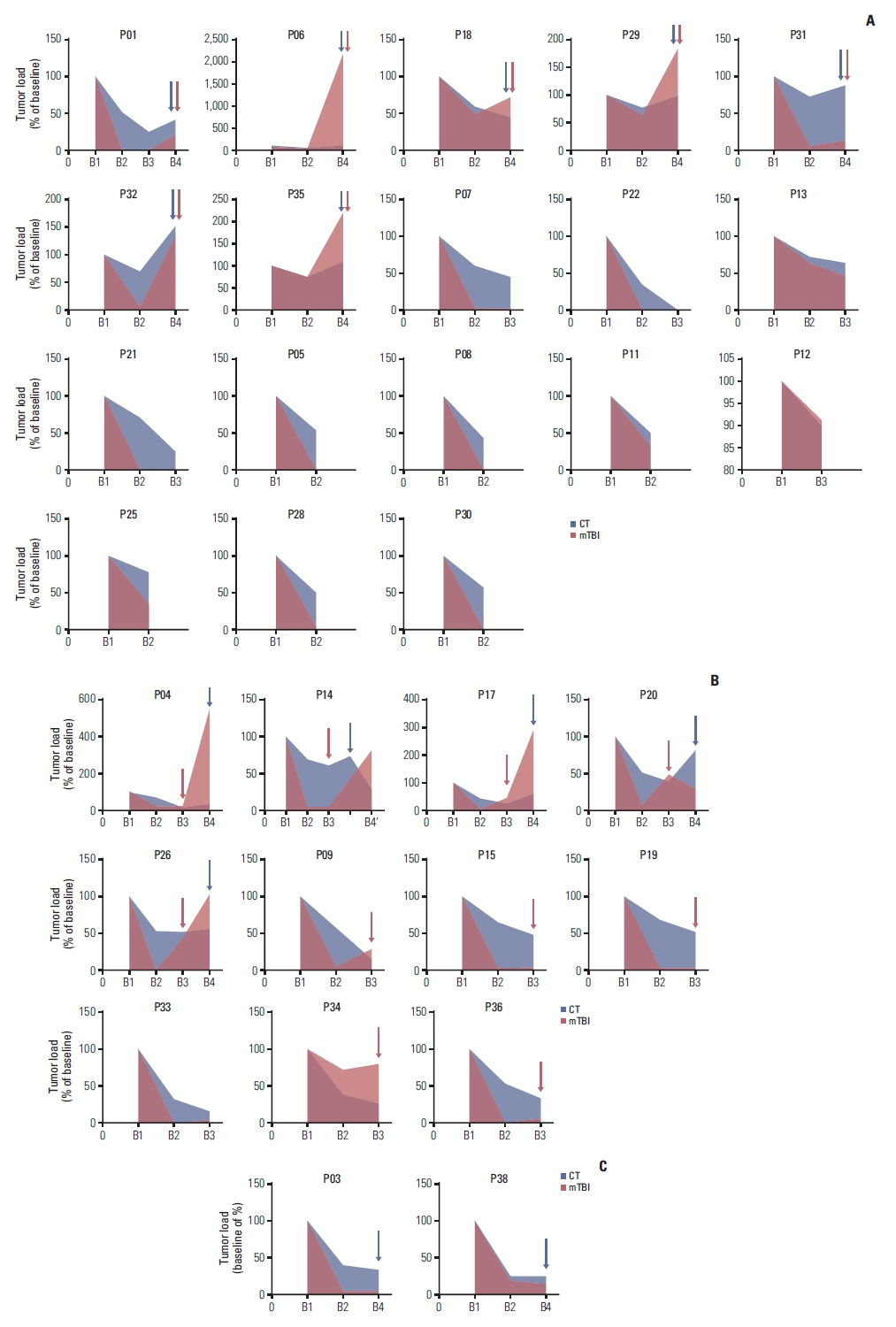
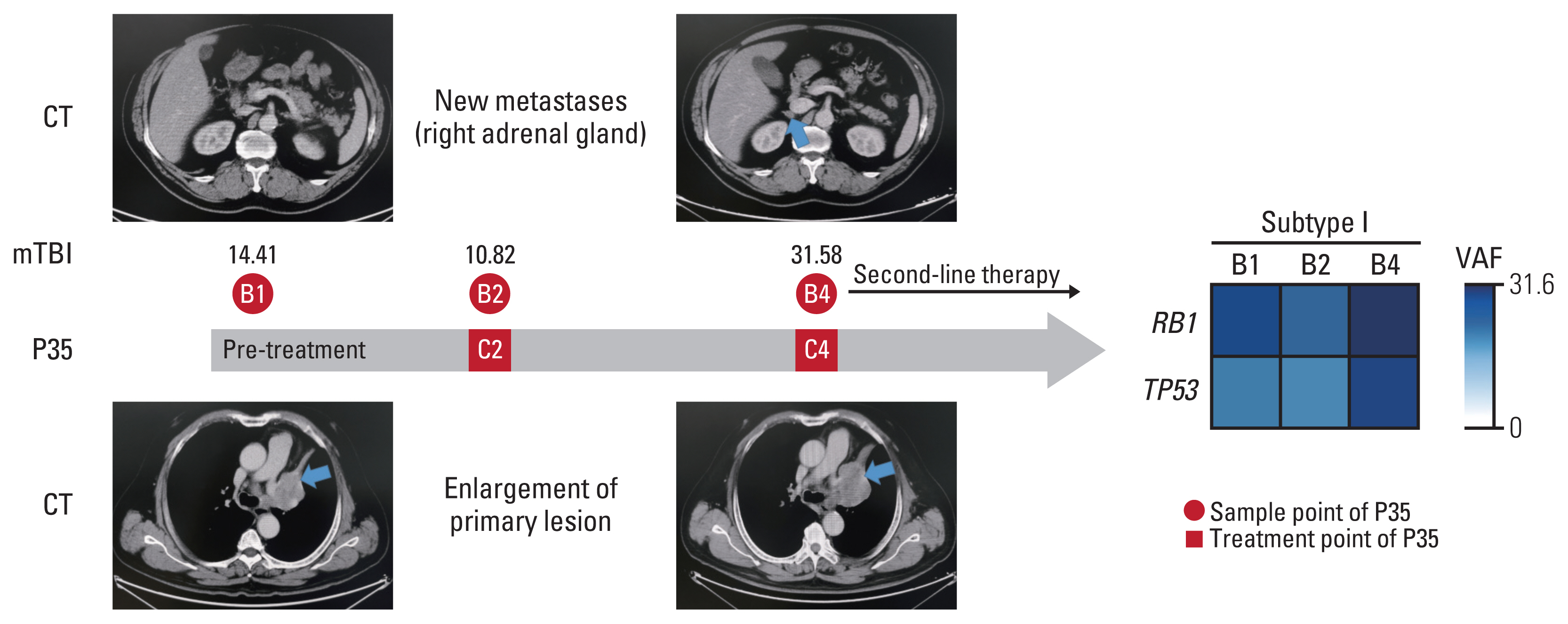
Tumor and longitudinal plasma ctDNA samples were analyzed by next-generation sequencing of 1,021 genes. PyClone was used to infer the molecular tumor burden index (mTBI). Pre-treatment tumor tissues [T1] and serial plasma samples were collected (pre-treatment [B1], after two [B2], six [B3] cycles of chemotherapy and at progression [B4]).
Results
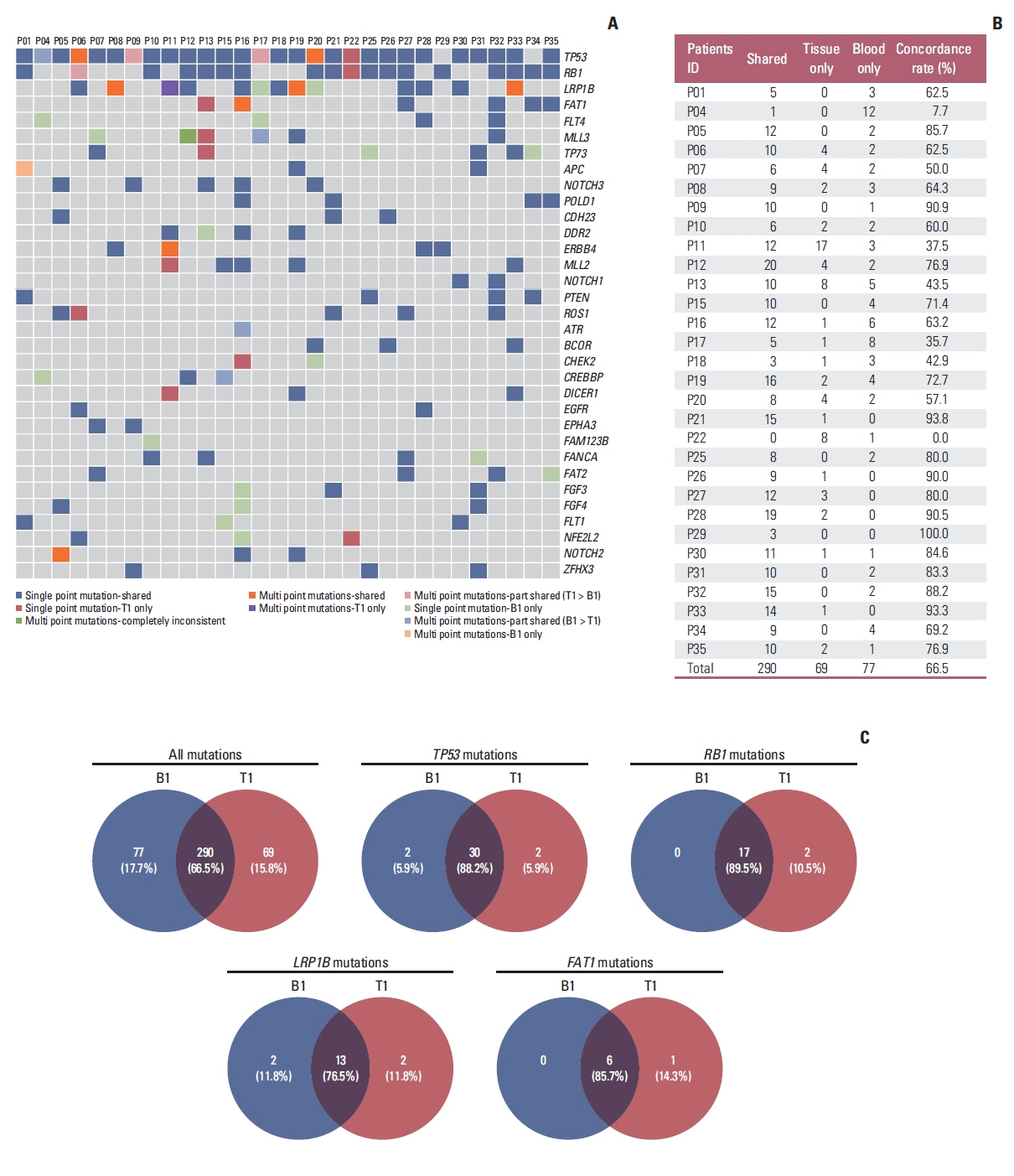
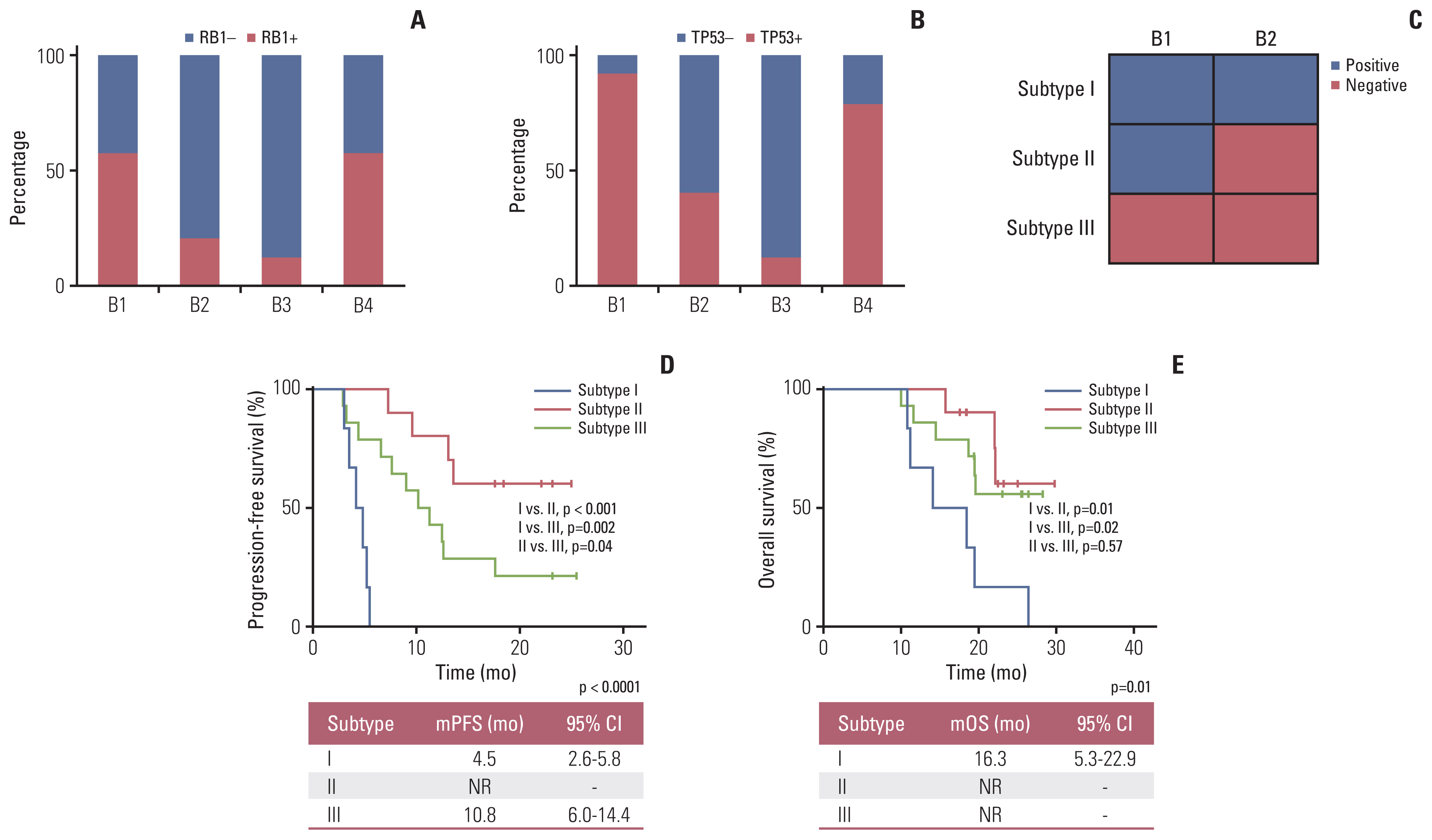
Overall concordance between T1 and B1 sequencing (n=30) was 66.5%, and 89.5% in the gene of RB1. A classification method was designed according to the changes of RB1 mutation, named as subtype Ⅰ (both positive at B1 and B2), subtype Ⅱ (positive at B1 but negative at B2), and subtype Ⅲ (both negative at B1 and B2). The median progressive-free survival for subtype Ⅰ patients (4.5 months [95%CI: 2.6-5.8]) was inferior to subtype Ⅱ (not reached, p<0.0001) and subtype Ⅲ (10.8 months [95%CI: 6.0-14.4], p=0.002). The median overall survival for subtype Ⅰ patients (16.3 months [95%CI: 5.3-22.9]) was inferior to subtype Ⅱ (not reached, p=0.01) and subtype Ⅲ (not reached, p=0.02). Patients with a mTBI dropped to zero at B2 had longer median overall survival (not reached vs. 19.5 months, p=0.01). The changes of mTBI from B4 to B1 were sensitive to predict new metastases, with a sensitivity of 100% and a specificity of 85.7%.
Conclusion
Monitoring ctDNA based RB1 mutation and mTBI provided a feasible tool to predict the prognosis of SCLC.
Keyword
Figure
Reference
-
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021; 71:209–49.
Article2. Govindan R, Page N, Morgensztern D, Read W, Tierney R, Vlahiotis A, et al. Changing epidemiology of small-cell lung cancer in the United States over the last 30 years: analysis of the surveillance, epidemiologic, and end results database. J Clin Oncol. 2006; 24:4539–44.
Article3. Elias AD. Small cell lung cancer: state-of-the-art therapy in 1996. Chest. 1997; 112(4 Suppl):251S–8S.4. Byers LA, Rudin CM. Small cell lung cancer: where do we go from here? Cancer. 2015; 121:664–72.
Article5. Lad T, Piantadosi S, Thomas P, Payne D, Ruckdeschel J, Giaccone G. A prospective randomized trial to determine the benefit of surgical resection of residual disease following response of small cell lung cancer to combination chemotherapy. Chest. 1994; 106(6 Suppl):320S–3S.
Article6. Yang CJ, Chan DY, Shah SA, Yerokun BA, Wang XF, D’Amico TA, et al. Long-term survival after surgery compared with concurrent chemoradiation for node-negative small cell lung cancer. Ann Surg. 2018; 268:1105–12.
Article7. Tjong MC, Mak DY, Shahi J, Li GJ, Chen H, Louie AV. Current management and progress in radiotherapy for small cell lung cancer. Front Oncol. 2020; 10:1146.
Article8. Horn L, Mansfield AS, Szczesna A, Havel L, Krzakowski M, Hochmair MJ, et al. First-line atezolizumab plus chemotherapy in extensive-stage small-cell lung cancer. N Engl J Med. 2018; 379:2220–9.
Article9. Paz-Ares L, Dvorkin M, Chen Y, Reinmuth N, Hotta K, Trukhin D, et al. Durvalumab plus platinum-etoposide versus platinum-etoposide in first-line treatment of extensive-stage small-cell lung cancer (CASPIAN): a randomised, controlled, open-label, phase 3 trial. Lancet. 2019; 394:1929–39.10. Ireland AS, Micinski AM, Kastner DW, Guo B, Wait SJ, Spain-hower KB, et al. MYC drives temporal evolution of small cell lung cancer subtypes by reprogramming neuroendocrine fate. Cancer Cell. 2020; 38:60–78.
Article11. Rudin CM, Poirier JT, Byers LA, Dive C, Dowlati A, George J, et al. Molecular subtypes of small cell lung cancer: a synthesis of human and mouse model data. Nat Rev Cancer. 2019; 19:289–97.
Article12. Yi X, Ma J, Guan Y, Chen R, Yang L, Xia X. The feasibility of using mutation detection in ctDNA to assess tumor dynamics. Int J Cancer. 2017; 140:2642–7.
Article13. Wang Y, Zhao C, Chang L, Jia R, Liu R, Zhang Y, et al. Circulating tumor DNA analyses predict progressive disease and indicate trastuzumab-resistant mechanism in advanced gastric cancer. EBioMedicine. 2019; 43:261–9.
Article14. Siravegna G, Marsoni S, Siena S, Bardelli A. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol. 2017; 14:531–48.
Article15. Gerwing M, Herrmann K, Helfen A, Schliemann C, Berdel WE, Eisenblatter M, et al. The beginning of the end for conventional RECIST: novel therapies require novel imaging approaches. Nat Rev Clin Oncol. 2019; 16:442–58.
Article16. Yu Q, Huang S, Wu Z, Zheng J, Chen X, Nie L. Label-free visualization of early cancer hepatic micrometastasis and intra-operative image-guided surgery by photoacoustic imaging. J Nucl Med. 2020; 61:1079–85.
Article17. Zhou L, Zhang M, Li R, Xue J, Lu Y. Pseudoprogression and hyperprogression in lung cancer: a comprehensive review of literature. J Cancer Res Clin Oncol. 2020; 146:3269–79.
Article18. Ma F, Guan Y, Yi Z, Chang L, Li Q, Chen S, et al. Assessing tumor heterogeneity using ctDNA to predict and monitor therapeutic response in metastatic breast cancer. Int J Cancer. 2020; 146:1359–68.
Article19. Nong J, Gong Y, Guan Y, Yi X, Yi Y, Chang L, et al. Circulating tumor DNA analysis depicts subclonal architecture and genomic evolution of small cell lung cancer. Nat Commun. 2018; 9:3114.
Article20. Mohan S, Foy V, Ayub M, Leong HS, Schofield P, Sahoo S, et al. Profiling of circulating free DNA using targeted and genome-wide sequencing in patients with SCLC. J Thorac Oncol. 2020; 15:216–30.
Article21. Almodovar K, Iams WT, Meador CB, Zhao Z, York S, Horn L, et al. Longitudinal cell-free DNA analysis in patients with small cell lung cancer reveals dynamic insights into treatment efficacy and disease relapse. J Thorac Oncol. 2018; 13:112–23.
Article22. George J, Lim JS, Jang SJ, Cun Y, Ozretic L, Kong G, et al. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015; 524:47–53.23. Roth A, Khattra J, Yap D, Wan A, Laks E, Biele J, et al. PyClone: statistical inference of clonal population structure in cancer. Nat Methods. 2014; 11:396–8.
Article24. Yi Z, Ma F, Rong G, Liu B, Guan Y, Li J, et al. The molecular tumor burden index as a response evaluation criterion in breast cancer. Signal Transduct Target Ther. 2021; 6:251.
Article25. Dawson SJ, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin SF, et al. Analysis of circulating tumor DNA to monitor meta-static breast cancer. N Engl J Med. 2013; 368:1199–209.
Article26. Babayan A, Pantel K. Advances in liquid biopsy approaches for early detection and monitoring of cancer. Genome Med. 2018; 10:21.
Article27. Rudin CM, Brambilla E, Faivre-Finn C, Sage J. Small-cell lung cancer. Nat Rev Dis Primers. 2021; 7:3.
Article28. Zhou CC, Imamura F, Cheng Y, Okamoto I, Cho BC, Lin MC, et al. Early clearance of plasma EGFR mutations as a predictor of response to osimertinib and comparator EGFR-TKIs in the FLAURA trial. J Clin Oncol. 2019; 37(15 Suppl):9020.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Clinical Application of Circulating Tumor DNA Analysis
- Circulating Cell-free Tumor Nucleic Acids in Gastric Cancer
- Circulating Tumor Cell and Cell-free Circulating Tumor DNA in Lung Cancer
- Revolutionizing Non–Small Cell Lung Cancer Diagnosis: Ultra-High-Sensitive ctDNA Analysis for Detecting Hotspot Mutations with Long-term Stored Plasma
- Current status and future perspectives of liquid biopsy in non-small cell lung cancer