Cancer Res Treat.
2022 Apr;54(2):424-433. 10.4143/crt.2021.583.
Comparison of the Predictive Power of a Combination versus Individual Biomarker Testing in Non–Small Cell Lung Cancer Patients Treated with Immune Checkpoint Inhibitors
- Affiliations
-
- 1Department of Pathology, Seoul National University Bundang Hospital, Seongnam, Korea
- 2Department of Pathology, Seoul National University College of Medicine, Seoul, Korea
- 3Department of Internal Medicine, Seoul National University Bundang Hospital, Seongnam, Korea
- KMID: 2528212
- DOI: http://doi.org/10.4143/crt.2021.583
Abstract
- Purpose
Since tumor mutational burden (TMB) and gene expression profiling (GEP) have complementary effects, they may have improved predictive power when used in combination. Here, we investigated the ability of TMB and GEP to predict the immunotherapy response in patients with non–small cell lung cancer (NSCLC) and assessed if this combination can improve predictive power compared to that when used individually.
Materials and Methods
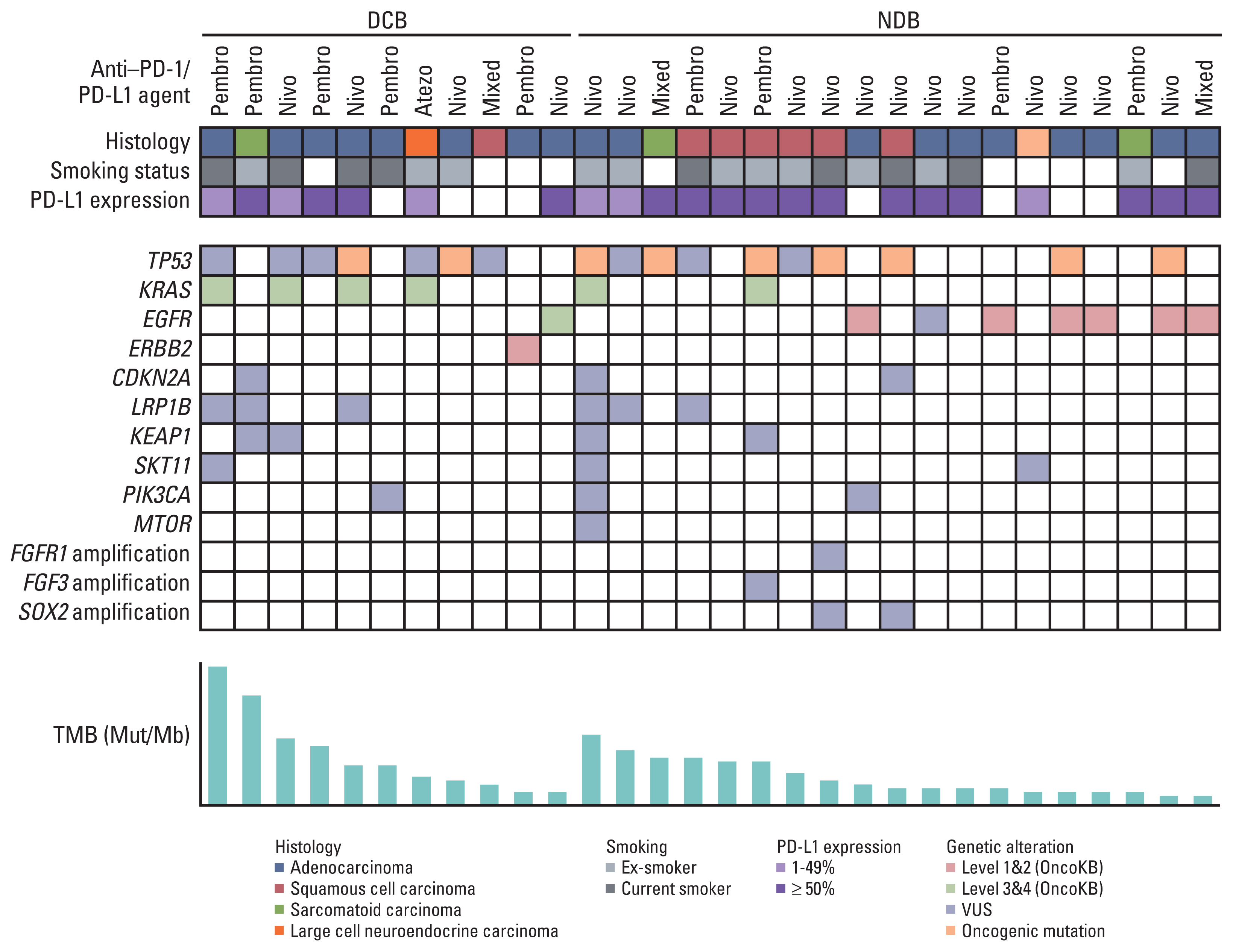
This retrospective cohort study included 30 patients with NSCLC who received immune checkpoint inhibitors (ICI) therapy at the Seoul National University Bundang Hospital. programmed cell death-ligand-1 (PD-L1) protein expression was assessed using immunohistochemistry, and TMB was measured by targeted deep sequencing. Gene expression was determined using NanoString nCounter analysis for the PanCancer IO360 panel, and enrichment analysis were performed.
Results
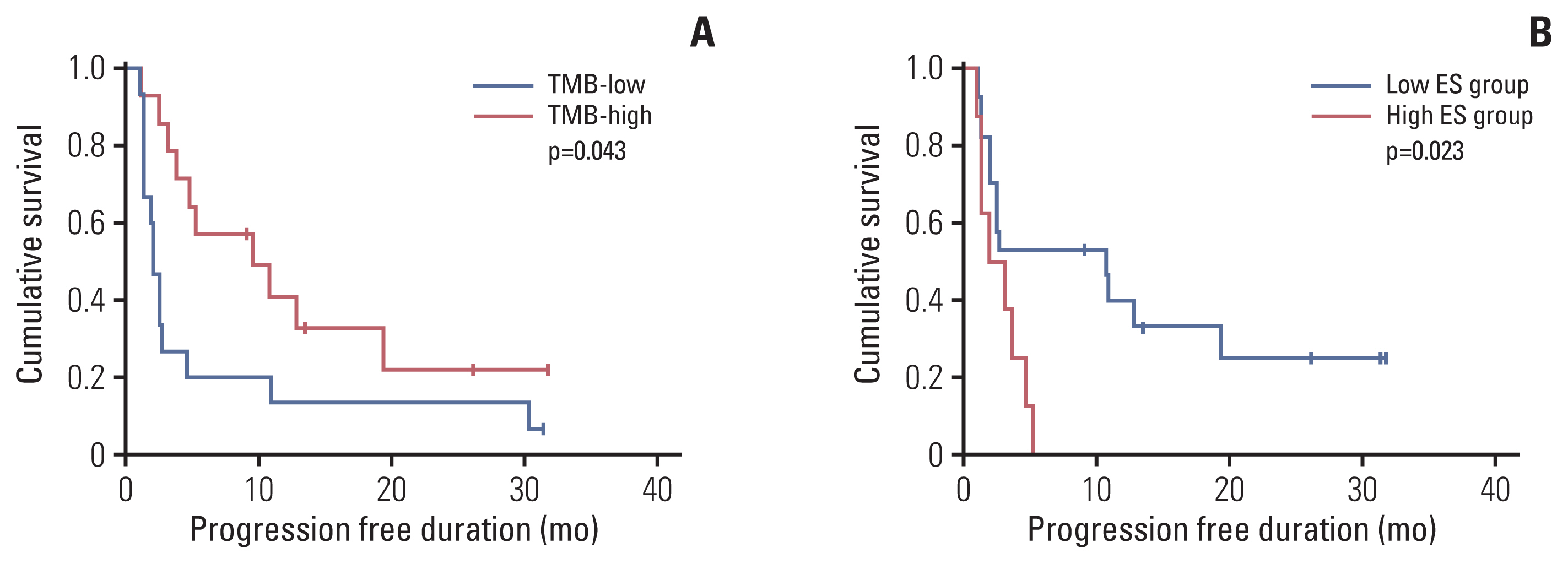
Eleven patients (36.7%) showed a durable clinical benefit (DCB), whereas 19 (63.3%) showed no durable benefit (NDB). TMB and enrichment scores (ES) showed significant differences between the DCB and NDB groups (p=0.044 and p=0.017, respectively); however, no significant correlations were observed among TMB, ES, and PD-L1. ES was the best single biomarker for predicting DCB (area under the curve [AUC], 0.794), followed by TMB (AUC, 0.679) and PD-L1 (AUC, 0.622). TMB and ES showed the highest AUC (0.837) among other combinations (AUC [TMB and PD-L1], 0.777; AUC [PD-L1 and ES], 0.763) and was similar to that of all biomarkers used together (0.832).
Conclusion
The combination of TMB and ES may be an effective predictive tool to identify patients with NSCLC patients who would possibly benefit from ICI therapies.
Keyword
Figure
Reference
-
References
1. Brahmer J, Reckamp KL, Baas P, Crino L, Eberhardt WE, Poddubskaya E, et al. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N Engl J Med. 2015; 373:123–35.
Article2. Borghaei H, Paz-Ares L, Horn L, Spigel DR, Steins M, Ready NE, et al. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N Engl J Med. 2015; 373:1627–39.
Article3. Herbst RS, Baas P, Kim DW, Felip E, Perez-Gracia JL, Han JY, et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet. 2016; 387:1540–50.
Article4. Rittmeyer A, Barlesi F, Waterkamp D, Park K, Ciardiello F, von Pawel J, et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): a phase 3, open-label, multicentre randomised controlled trial. Lancet. 2017; 389:255–65.
Article5. Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Cancer immunology: mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015; 348:124–8.
Article6. Yarchoan M, Hopkins A, Jaffee EM. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med. 2017; 377:2500–1.
Article7. Hellmann MD, Callahan MK, Awad MM, Calvo E, Ascierto PA, Atmaca A, et al. Tumor mutational burden and efficacy of nivolumab monotherapy and in combination with ipilimumab in small-cell lung cancer. Cancer Cell. 2018; 33:853–61.
Article8. Goodman AM, Kato S, Bazhenova L, Patel SP, Frampton GM, Miller V, et al. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther. 2017; 16:2598–608.
Article9. Hellmann MD, Paz-Ares L, Bernabe Caro R, Zurawski B, Kim SW, Carcereny Costa E, et al. Nivolumab plus ipilimumab in advanced non-small-cell lung cancer. N Engl J Med. 2019; 381:2020–31.
Article10. Auslander N, Zhang G, Lee JS, Frederick DT, Miao B, Moll T, et al. Robust prediction of response to immune checkpoint blockade therapy in metastatic melanoma. Nat Med. 2018; 24:1545–9.
Article11. Jiang P, Gu S, Pan D, Fu J, Sahu A, Hu X, et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med. 2018; 24:1550–8.
Article12. Yu Y, Zeng D, Ou Q, Liu S, Li A, Chen Y, et al. Association of survival and immune-related biomarkers with immunotherapy in patients with non-small cell lung xancer: a meta-analysis and individual patient-level analysis. JAMA Netw Open. 2019; 2:e196879.13. Lee JS, Ruppin E. Multiomics prediction of response rates to therapies to inhibit programmed cell death 1 and programmed cell death 1 ligand 1. JAMA Oncol. 2019; 5:1614–8.
Article14. Lu S, Stein JE, Rimm DL, Wang DW, Bell JM, Johnson DB, et al. Comparison of biomarker modalities for predicting response to PD-1/PD-L1 checkpoint blockade: a systematic review and meta-analysis. JAMA Oncol. 2019; 5:1195–204.
Article15. Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009; 45:228–47.
Article16. Hernandez-Prera JC, Valderrabano P, Creed JH, de la Iglesia JV, Slebos RJC, Centeno BA, et al. Molecular determinants of thyroid nodules with indeterminate cytology and RAS mutations. Thyroid. 2021; 31:36–49.
Article17. Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006; 38:500–1.
Article18. Kim H, Kwon HJ, Han YB, Park SY, Kim ES, Kim SH, et al. Increased CD3+ T cells with a low FOXP3+/CD8+ T cell ratio can predict anti-PD-1 therapeutic response in non-small cell lung cancer patients. Mod Pathol. 2019; 32:367–75.
Article19. Ott PA, Bang YJ, Piha-Paul SA, Razak ARA, Bennouna J, Soria JC, et al. T-cell-inflamed gene-expression profile, programmed death ligand 1 expression, and tumor mutational burden predict efficacy in patients treated with pembrolizumab across 20 cancers: KEYNOTE-028. J Clin Oncol. 2019; 37:318–27.
Article20. Feng YY, Griffith OL, Griffith M. Clinical implications of neoepitope landscapes for adult and pediatric cancers. Genome Med. 2017; 9:77.
Article21. Turajlic S, Litchfield K, Xu H, Rosenthal R, McGranahan N, Reading JL, et al. Insertion-and-deletion-derived tumour-specific neoantigens and the immunogenic phenotype: a pan-cancer analysis. Lancet Oncol. 2017; 18:1009–21.
Article22. McGrail DJ, Pilie PG, Rashid NU, Voorwerk L, Slagter M, Kok M, et al. High tumor mutation burden fails to predict immune checkpoint blockade response across all cancer types. Ann Oncol. 2021; 32:661–72.
Article23. Dees S, Ganesan R, Singh S, Grewal IS. Regulatory T cell targeting in cancer: emerging strategies in immunotherapy. Eur J Immunol. 2021; 51:280–91.
Article24. Paluskievicz CM, Cao X, Abdi R, Zheng P, Liu Y, Bromberg JS. T regulatory cells and priming the suppressive tumor microenvironment. Front Immunol. 2019; 10:2453.
Article25. Yarchoan M, Albacker LA, Hopkins AC, Montesion M, Murugesan K, Vithayathil TT, et al. PD-L1 expression and tumor mutational burden are independent biomarkers in most cancers. JCI Insight. 2019; 4:e126908.
Article26. Gainor JF, Shaw AT, Sequist LV, Fu X, Azzoli CG, Piotrowska Z, et al. EGFR mutations and ALK rearrangements are associated with low response rtes to PD-1 pathway blockade in non-small cell lung cancer: a retrospective analysis. Clin Cancer Res. 2016; 22:4585–93.27. Dong ZY, Zhong WZ, Zhang XC, Su J, Xie Z, Liu SY, et al. Potential predictive value of TP53 and KRAS mutation status for response to PD-1 blockade immunotherapy in lung adenocarcinoma. Clin Cancer Res. 2017; 23:3012–24.28. Skoulidis F, Goldberg ME, Greenawalt DM, Hellmann MD, Awad MM, Gainor JF, et al. STK11/LKB1 mutations and PD-1 inhibitor resistance in KRAS-mutant lung adenocarcinoma. Cancer Discov. 2018; 8:822–35.29. Papillon-Cavanagh S, Doshi P, Dobrin R, Szustakowski J, Walsh AM. STK11 and KEAP1 mutations as prognostic biomarkers in an observational real-world lung adenocarcinoma cohort. ESMO Open. 2020; 5:e000706.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- PD-L1 Testing in Non-small Cell Lung Cancer: Past, Present, and Future
- Exploring histological predictive biomarkers for immune checkpoint inhibitor therapy response in non–small cell lung cancer
- Current status of cancer immunotherapy with immune checkpoint inhibitors
- Tumor Immunology and Immune Checkpoint Inhibitors in Non-Small Cell Lung Cancer
- Immunotherapy for Non-small-cell Lung Cancer: Current Status and Future Obstacles