Int J Stem Cells.
2021 Feb;14(1):85-93. 10.15283/ijsc20073.
Dysregulated Dermal Mesenchymal Stem Cell Proliferation and Differentiation Interfered by Glucose Metabolism in Psoriasis
- Affiliations
-
- 1Shanxi Key Laboratory of Stem Cell for Immunological Dermatosis, Institute of Dermatology, Taiyuan City Centre Hospital of Shanxi Medical University, Taiyuan, China
- KMID: 2513085
- DOI: http://doi.org/10.15283/ijsc20073
Abstract
- Background and Objectives
Psoriasis is a chronic inflammatory skin disease, which the mechanisms behind its initiation and development are related to many factors. DMSCs (dermal mesenchymal stem cells) represent an important member of the skin microenvironment and play an important role in the surrounding environment and in neighbouring cells, but they are also affected by the microenvironment. We studied the glucose metabolism of DMSCs in psoriasis patients and a control group to reveal the relationship among glucose metabolism, cell proliferation activity,and VEC (vascular endothelial cell) differentiation in vitro, we demonstrated the biological activity and molecular mechanisms of DMSCs in psoriasis.
Methods and Results
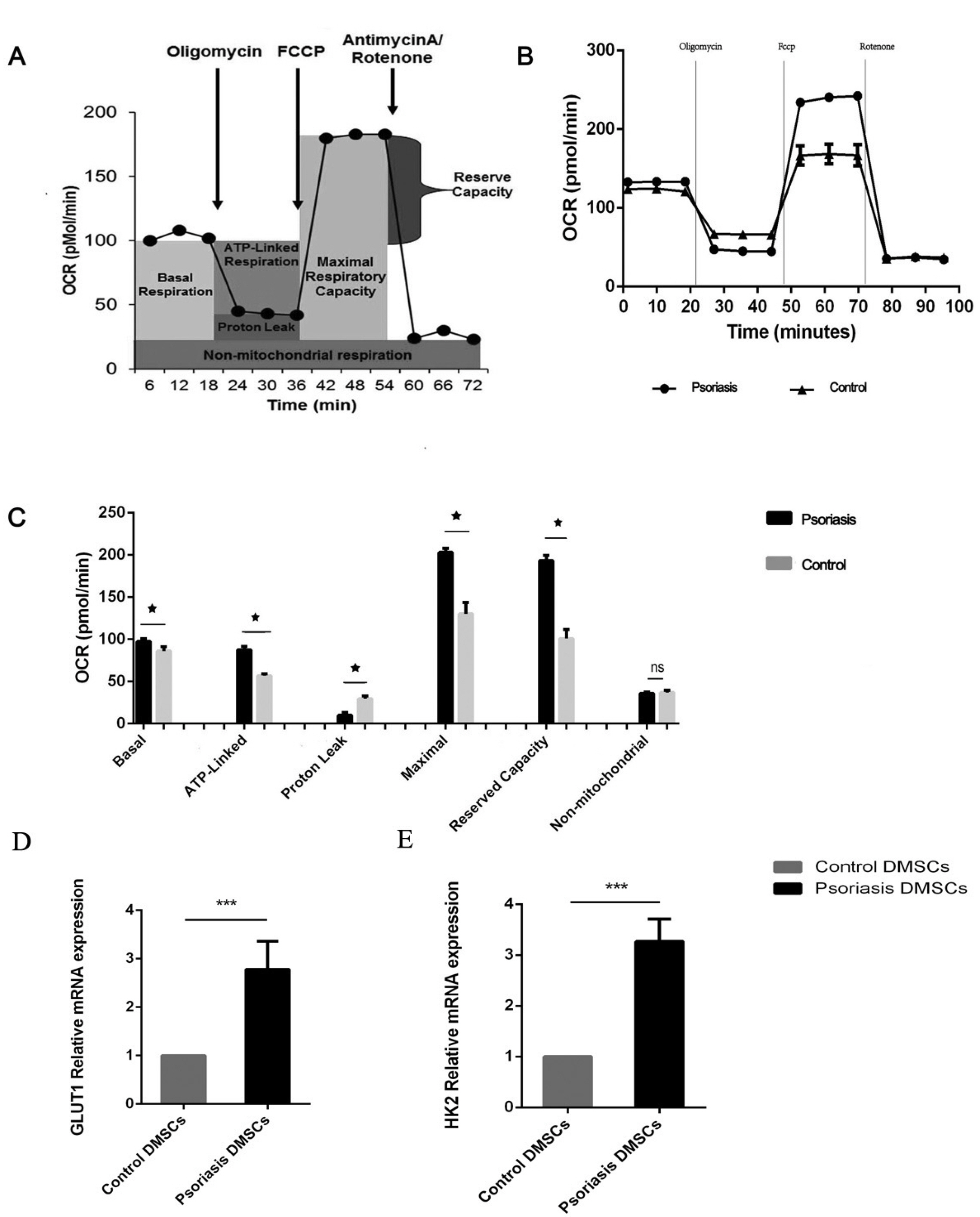
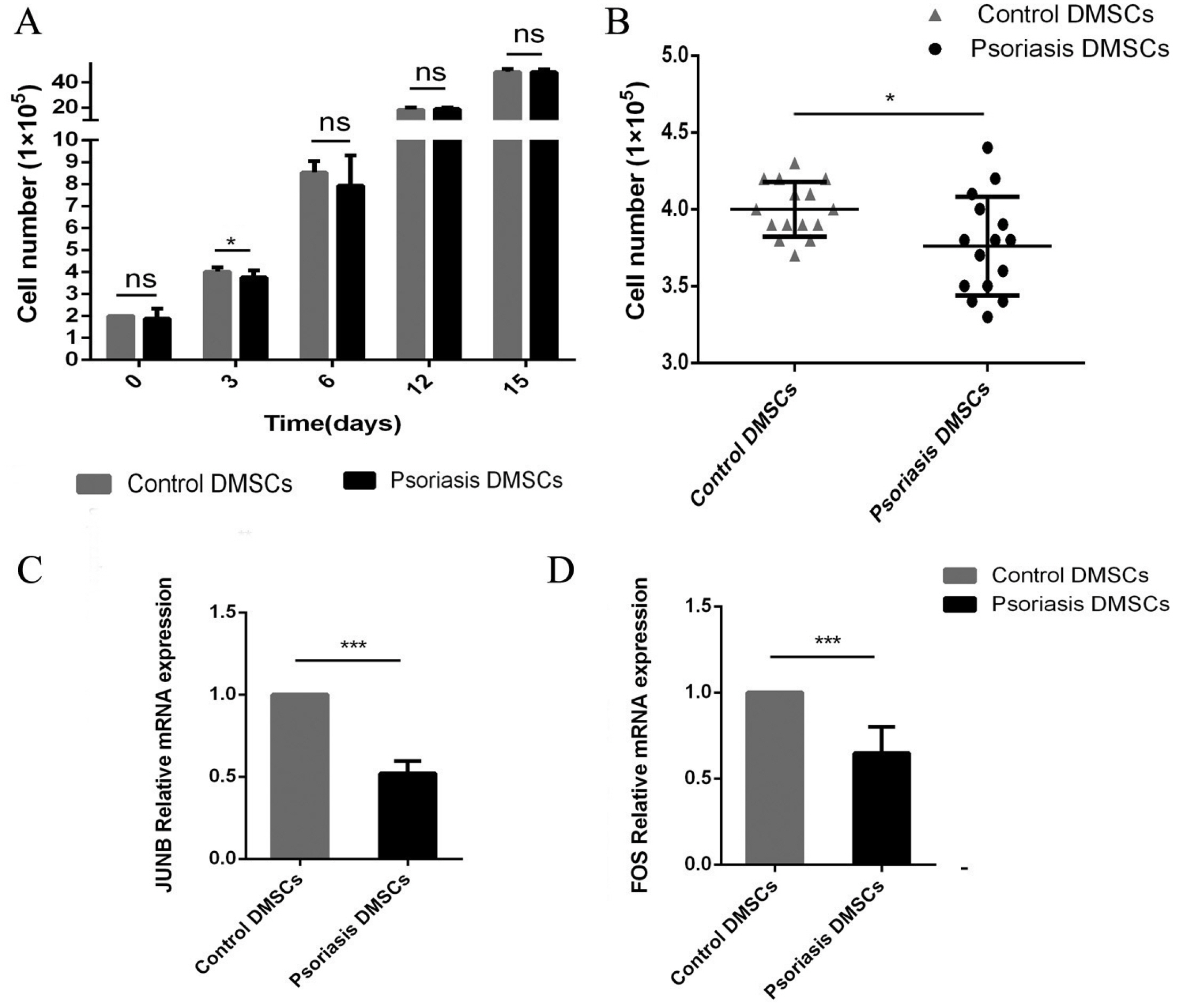
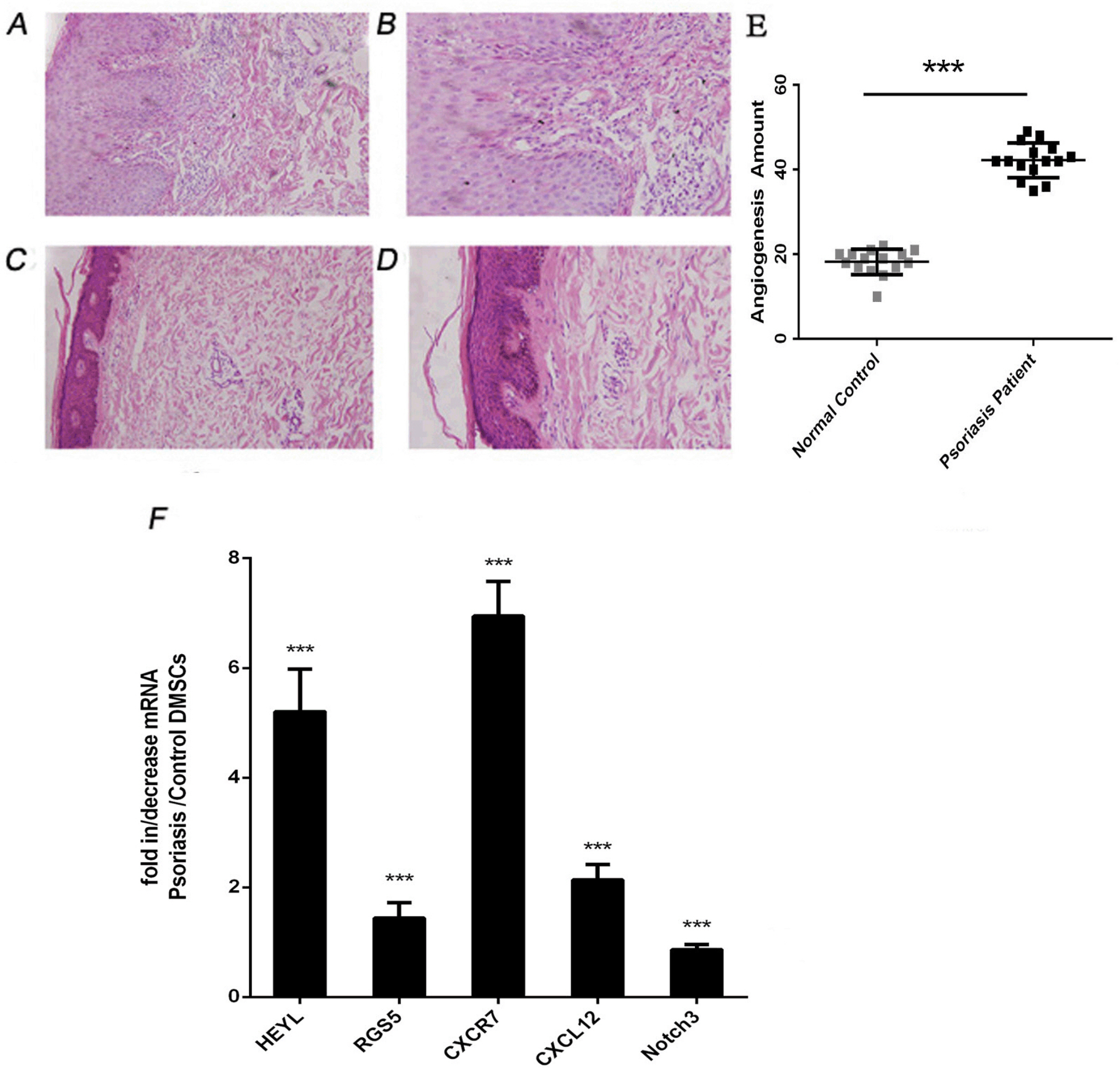
We found that the OCR of DMSCs in psoriatic lesions was higher than that in the control group, and mRNA of GLUT1 and HK2 were up-regulated compared with the control group. The proliferative activity of DMSCs in psoriasis was reduced at an early stage, and mRNA involved in proliferation, JUNB and FOS were expressed at lower levels than those in the control group. The number of blood vessels in psoriatic lesions was significantly higher than that in the control group (p<0.05), which the mRNA of VEC differentiation, CXCL12, CXCR7, HEYL and RGS5 tended to be increased in psoriatic lesions compared to the control group, in addition to Notch3.
Conclusions
We speculated that DMSCs affected local psoriatic blood vessels through glucose metabolism, and the differentiation of VECs, which resulted in the pathophysiological process of psoriasis.
Figure
Reference
-
References
1. Korman NJ. 2020; Management of psoriasis as a systemic disease: what is the evidence? Br J Dermatol. 182:840–848. DOI: 10.1111/bjd.18245. PMID: 31225638. PMCID: PMC7187293.
Article2. Crow JM. 2012; Psoriasis uncovered. Nature. 492:S50–S51. DOI: 10.1038/492S50a. PMID: 23254970.
Article3. Rapp SR, Feldman SR, Exum ML, Fleischer AB Jr, Reboussin DM. 1999; Psoriasis causes as much disability as other major medical diseases. J Am Acad Dermatol. 41(3 Pt 1):401–407. DOI: 10.1016/S0190-9622(99)70112-X. PMID: 31837740.
Article4. Ryan C, Kirby B. 2015; Psoriasis is a systemic disease with multiple cardiovascular and metabolic comorbidities. Dermatol Clin. 33:41–55. DOI: 10.1016/j.det.2014.09.004. PMID: 25412782.
Article5. Zhang Z, Zi Z, Lee EE, Zhao J, Contreras DC, South AP, Abel ED, Chong BF, Vandergriff T, Hosler GA, Scherer PE, Mettlen M, Rathmell JC, DeBerardinis RJ, Wang RC. 2018; Differential glucose requirement in skin homeostasis and injury identifies a therapeutic target for psoriasis. Nat Med. 24:617–627. DOI: 10.1038/s41591-018-0003-0. PMID: 29662201. PMCID: PMC6095711.
Article6. Vieira Paladino F, de Moraes Rodrigues J, da Silva A, Goldberg AC. 2019; The immunomodulatory potential of Wharton's jelly mesenchymal stem/stromal cells. Stem Cells Int. 2019:3548917. DOI: 10.1155/2019/3548917. PMID: 31281372. PMCID: PMC6594275.
Article7. Park IS, Chung PS, Ahn JC. 2015; Adipose-derived stromal cell cluster with light therapy enhance angiogenesis and skin wound healing in mice. Biochem Biophys Res Commun. 462:171–177. DOI: 10.1016/j.bbrc.2015.04.059. PMID: 25911320.
Article8. De Jesus MM, Santiago JS, Trinidad CV, See ME, Semon KR, Fernandez MO Jr, Chung FS. 2016; Autologous adipose-derived mesenchymal stromal cells for the treatment of psoriasis vulgaris and psoriatic arthritis: a case report. Cell Transplant. 25:2063–2069. DOI: 10.3727/096368916X691998. PMID: 27301844.
Article9. Chen H, Niu JW, Ning HM, Pan X, Li XB, Li Y, Wang DH, Hu LD, Sheng HX, Xu M, Zhang L, Zhang B. 2016; Treatment of psoriasis with mesenchymal stem cells. Am J Med. 129:e13–e14. DOI: 10.1016/j.amjmed.2015.11.001. PMID: 26582058.
Article10. Liu R, Yang Y, Yan X, Zhang K. 2013; Abnormalities in cytokine secretion from mesenchymal stem cells in psoriatic skin lesions. Eur J Dermatol. 23:600–607. DOI: 10.1684/ejd.2013.2149. PMID: 24135516.
Article11. Hou R, Yin G, An P, Wang C, Liu R, Yang Y, Yan X, Li J, Li X, Zhang K. 2013; DNA methylation of dermal MSCs in psoriasis: identification of epigenetically dysregulated genes. J Dermatol Sci. 72:103–109. DOI: 10.1016/j.jdermsci.2013.07.002. PMID: 23916410.
Article12. Li J, Xing J, Lu F, Chang W, Liang N, Li J, Wang Y, Li X, Zhao X, Hou R, Man M, Yin G, Li X, Zhang K. 2020; Psoriatic dermal-derived mesenchymal stem cells reduce keratinocyte junctions, and increase glycolysis. Acta Derm Venereol. 100:adv00122. DOI: 10.2340/00015555-3480. PMID: 32266413.
Article13. Cheng H, Qiu L, Zhang H, Cheng M, Li W, Zhao X, Liu K, Lei L, Ma J. 2011; Arsenic trioxide promotes senescence and regulates the balance of adipogenic and osteogenic differentiation in human mesenchymal stem cells. Acta Biochim Biophys Sin (Shanghai). 43:204–209. DOI: 10.1093/abbs/gmq130. PMID: 21257625.
Article14. Zhou L, Niu X, Liang J, Li J, Li J, Cheng Y, Meng Y, Wang Q, Yang X, Wang G, Shi Y, Dang E, Zhang K. 2018; Efficient differentiation of vascular endothelial cells from dermal-derived mesenchymal stem cells induced by endothelial cell lines conditioned medium. Acta Histochem. 120:734–740. DOI: 10.1016/j.acthis.2018.08.004. PMID: 30143315.
Article15. Mulukutla BC, Yongky A, Le T, Mashek DG, Hu WS. 2016; Regulation of glucose metabolism - a perspective from cell bioprocessing. Trends Biotechnol. 34:638–651. DOI: 10.1016/j.tibtech.2016.04.012. PMID: 27265890.
Article16. Friis NU, Hoffmann N, Gyldenlove M, Skov L, Vilsboll T, Knop FK, Storgaard H. 2019; Glucose metabolism in patients with psoriasis. Br J Dermatol. 180:264–271. DOI: 10.1111/bjd.17349. PMID: 30376181.
Article17. Li J, Zhou L, Liang J, Liu Y, Li J, Hou H, Hou R, Niu X, Li J, Liu R, Zhao X, Meng Y, Yang X, Wang G, Shi Y, Dang E, Zhang K. 2018; Psoriatic mesenchymal stem cells demonstrate an enhanced ability to differentiate into vascular endothelial cells. Eur J Dermatol. 28:688–690. DOI: 10.1684/ejd.2018.3344. PMID: 29941414.18. Hodeib AA, Neinaa YME, Zakaria SS, Alshenawy HA. 2018; Glucose transporter-1 (GLUT-1) expression in psoriasis: correlation with disease severity. Int J Dermatol. 57:943–951. DOI: 10.1111/ijd.14037. PMID: 29797802.
Article19. Roberts DJ, Miyamoto S. 2015; Hexokinase II integrates energy metabolism and cellular protection: akting on mitochondria and TORCing to autophagy. Cell Death Differ. 22:248–257. DOI: 10.1038/cdd.2014.173. PMID: 25323588. PMCID: PMC4291497.
Article20. Hou R, Yan H, Niu X, Chang W, An P, Wang C, Yang Y, Yan X, Li J, Liu R, Li X, Zhang K. 2014; Gene expression profile of dermal mesenchymal stem cells from patients with psoriasis. J Eur Acad Dermatol Venereol. 28:1782–1791. DOI: 10.1111/jdv.12420. PMID: 24593802.
Article21. Brownstein MH. 1966; Psoriasis and diabetes mellitus. Arch Dermatol. 93:654–655. DOI: 10.1001/archderm.1966.01600240020003. PMID: 5932149.
Article22. Makuch S, Woźniak M, Krawczyk M, Pastuch-Gawołek G, Szeja W, Agrawal S. 2020; Glycoconjugation as a promising treatment strategy for psoriasis. J Pharmacol Exp Ther. 373:204–212. DOI: 10.1124/jpet.119.263657. PMID: 32156758.
Article23. Choi SY, Heo MJ, Lee C, Choi YM, An IS, Bae S, An S, Jung JH. 2020; 2-deoxy-d-glucose Ameliorates animal models of dermatitis. Biomedicines. 8:20. DOI: 10.3390/biomedicines8020020. PMID: 31991554. PMCID: PMC7167934.
Article24. Gill KS, Fernandes P, O'Donovan TR, McKenna SL, Doddakula KK, Power DG, Soden DM, Forde PF. 2016; Glycolysis inhibition as a cancer treatment and its role in an anti-tumour immune response. Biochim Biophys Acta. 1866:87–105. DOI: 10.1016/j.bbcan.2016.06.005. PMID: 27373814.
Article25. Alfonso-Gonzalez C, Riesgo-Escovar JR. 2018; Fos metamorphoses: lessons from mutants in model organisms. Mech Dev. 154:73–81. DOI: 10.1016/j.mod.2018.05.006. PMID: 29753813.
Article26. Zhao L, Huang J, Guo R, Wang Y, Chen D, Xing L. 2010; Smurf1 inhibits mesenchymal stem cell proliferation and differentiation into osteoblasts through JunB degradation. J Bone Miner Res. 25:1246–1256. DOI: 10.1002/jbmr.28. PMID: 20200942. PMCID: PMC3153132.
Article27. Chiu R, Boyle WJ, Meek J, Smeal T, Hunter T, Karin M. 1988; The c-Fos protein interacts with c-Jun/AP-1 to stimulate transcription of AP-1 responsive genes. Cell. 54:541–552. DOI: 10.1016/0092-8674(88)90076-1. PMID: 3135940.
Article28. Fan F, Bashari MH, Morelli E, Tonon G, Malvestiti S, Vallet S, Jarahian M, Seckinger A, Hose D, Bakiri L, Sun C, Hu Y, Ball CR, Glimm H, Sattler M, Goldschmidt H, Wagner EF, Tassone P, Jaeger D, Podar K. 2017; The AP-1 transcription factor JunB is essential for multiple myeloma cell proliferation and drug resistance in the bone marrow microenvironment. Leukemia. 31:1570–1581. DOI: 10.1038/leu.2016.358. PMID: 27890927.
Article29. Martins GA, Cimmino L, Liao J, Magnusdottir E, Calame K. 2008; Blimp-1 directly represses Il2 and the Il2 activator Fos, attenuating T cell proliferation and survival. J Exp Med. 205:1959–1965. DOI: 10.1084/jem.20080526. PMID: 18725523. PMCID: PMC2526191.
Article30. He Z, Jiang J, Kokkinaki M, Golestaneh N, Hofmann MC, Dym M. 2008; Gdnf upregulates c-Fos transcription via the Ras/Erk1/2 pathway to promote mouse spermatogonial stem cell proliferation. Stem Cells. 26:266–278. DOI: 10.1634/stemcells.2007-0436. PMID: 17962702. PMCID: PMC2905627.
Article31. Salem HK, Thiemermann C. 2010; Mesenchymal stromal cells: current understanding and clinical status. Stem Cells. 28:585–596. DOI: 10.1002/stem.269. PMID: 19967788. PMCID: PMC2962904.
Article32. Virgintino D, Errede M, Rizzi M, Girolamo F, Strippoli M, Wälchli T, Robertson D, Frei K, Roncali L. 2013; The CXCL12/CXCR4/CXCR7 ligand-receptor system regulates neuro-glio-vascular interactions and vessel growth during human brain development. J Inherit Metab Dis. 36:455–466. DOI: 10.1007/s10545-012-9574-y. PMID: 23344887.
Article33. Weng Y, Lou J, Liu X, Lin S, Xu C, Du C, Tang L. 2019; Effects of high glucose on proliferation and function of circulating fibrocytes: involvement of CXCR4/SDF‑1 axis. Int J Mol Med. 44:927–938. DOI: 10.3892/ijmm.2019.4260. PMID: 31257476. PMCID: PMC6657976.
Article34. Hurst JH, Hooks SB. 2009; Regulator of G-protein signaling (RGS) proteins in cancer biology. Biochem Pharmacol. 78:1289–1297. DOI: 10.1016/j.bcp.2009.06.028. PMID: 19559677.
Article35. Yen WC, Fischer MM, Axelrod F, Bond C, Cain J, Cancilla B, Henner WR, Meisner R, Sato A, Shah J, Tang T, Wallace B, Wang M, Zhang C, Kapoun AM, Lewicki J, Gurney A, Hoey T. 2015; Targeting Notch signaling with a Notch2/Notch3 antagonist (tarextumab) inhibits tumor growth and decreases tumor-initiating cell frequency. Clin Cancer Res. 21:2084–2095. DOI: 10.1158/1078-0432.CCR-14-2808. PMID: 25934888.
Article36. Lee SY, Long F. 2018; Notch signaling suppresses glucose metabolism in mesenchymal progenitors to restrict osteoblast differentiation. J Clin Invest. 128:5573–5586. DOI: 10.1172/JCI96221. PMID: 30284985. PMCID: PMC6264656.
Article37. Fischer A, Steidl C, Wagner TU, Lang E, Jakob PM, Friedl P, Knobeloch KP, Gessler M. 2007; Combined loss of Hey1 and HeyL causes congenital heart defects because of impaired epithelial to mesenchymal transition. Circ Res. 100:856–863. DOI: 10.1161/01.RES.0000260913.95642.3b. PMID: 17303760.
Article38. Eelen G, de Zeeuw P, Simons M, Carmeliet P. 2015; Endothelial cell metabolism in normal and diseased vasculature. Circ Res. 116:1231–1244. DOI: 10.1161/CIRCRESAHA.116.302855. PMID: 25814684. PMCID: PMC4380230.
Article39. Vigo T, La Rocca C, Faicchia D, Procaccini C, Ruggieri M, Salvetti M, Centonze D, Matarese G, Uccelli A. 2019; IFNβ enhances mesenchymal stromal (Stem) cells immunomodulatory function through STAT1-3 activation and mTOR-associated promotion of glucose metabolism. Cell Death Dis. 10:85. DOI: 10.1038/s41419-019-1336-4. PMID: 30692524. PMCID: PMC6349843.
Article40. Zhang H, Badur MG, Divakaruni AS, Parker SJ, Jäger C, Hiller K, Murphy AN, Metallo CM. 2016; Distinct metabolic states can support self-renewal and lipogenesis in human pluripotent stem cells under different culture conditions. Cell Rep. 16:1536–1547. DOI: 10.1016/j.celrep.2016.06.102. PMID: 27477285. PMCID: PMC4981511.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- The Role of Histone Acetylation in Mesenchymal Stem Cell Differentiation
- A study of growth factors, chondrogenic differentiation of mesenchymal stem cells and cell response by needle size differences in vitro
- Stimulation of Chondrogenic Differentiation of Mesenchymal Stem Cells
- Psoriatic Serum Induce an Abnormal Inflammatory Phenotype and a Decreased Immunosuppressive Function of Mesenchymal Stem Cells
- Dynamic changes of gangliosides expression during the differentiation of embryonic and mesenchymal stem cells into neural cells