Clin Exp Vaccine Res.
2020 Jan;9(1):15-25. 10.7774/cevr.2020.9.1.15.
Reverse vaccinology approach for the identification and characterization of outer membrane proteins of Shigella flexneri as potential cellular- and antibody-dependent vaccine candidates
- Affiliations
-
- 1Institute for Research in Molecular Medicine, Universiti Sains Malaysia, Kubang Kerian, Malaysia. yee.leow@usm.my
- 2School of Medical Sciences, Universiti Sains Malaysia, Kubang Kerian, Malaysia.
- 3School of Health Sciences, Universiti Sains Malaysia, Kubang Kerian, Malaysia.
- 4Institute for Research in Molecular Medicine, Universiti Sains Malaysia, Gelugor, Malaysia.
- KMID: 2470834
- DOI: http://doi.org/10.7774/cevr.2020.9.1.15
Abstract
- PURPOSE
In the developing world, bacillary dysentery is one of the most common communicable diarrheal infections. There are approximately 169 million cases of shigellosis reported worldwide. The disease is transmitted by a group of Gram-negative intracellular enterobacteria known as Shigella flexneri, S. sonnei, S. dysenteriae, and S. boydii. Conventional treatment regimens for Shigella have been less effective due to the development of resistant strains against antibiotics. Therefore, an effective vaccine for the long term control of Shigella transmission is urgently needed.
MATERIALS AND METHODS
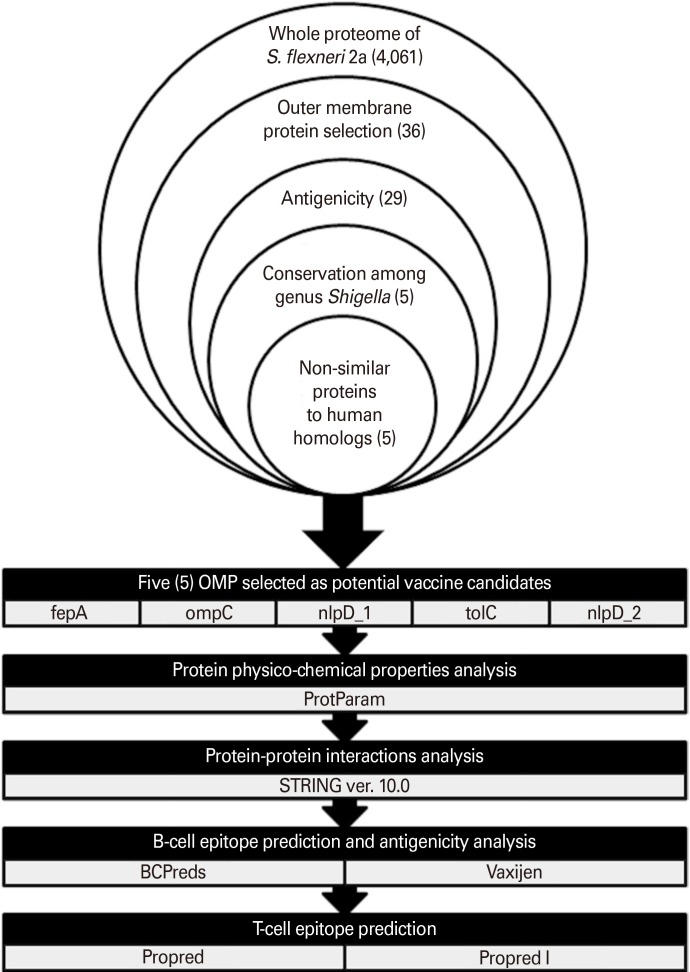
In this study, a reverse vaccinology approach was employed to identify most conserved and immunogenic outer membrane proteins (OMPs) of S. flexneri 2a.
RESULTS
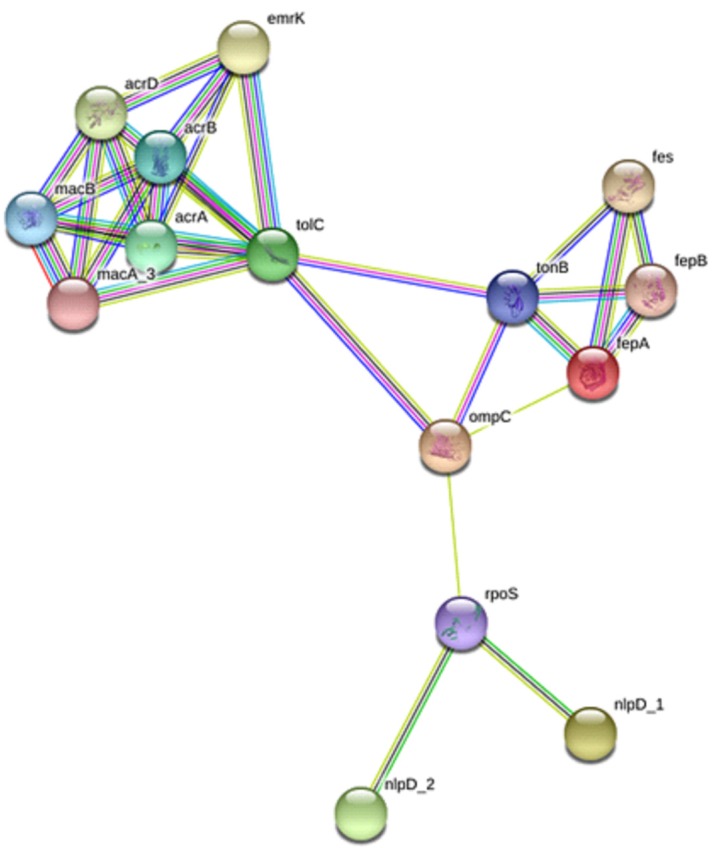
Five OMPs including fepA, ompC, nlpD_1, tolC, and nlpD_2 were identified as potential vaccine candidates. Protein-protein interactions analysis using STRING software (https://string-db.org/) revealed that five of these OMPs may potentially interact with other intracellular proteins which are involved in beta-lactam resistance pathway. B- and T-cell epitopes of the selected OMPs were predicted using BCPred as well as Propred I and Propred (http://crdd.osdd.net/raghava/propred/), respectively. Each of these OMPs contains regions which are capable to induce B- and T-cell immune responses.
CONCLUSION
Analysis acquired from this study showed that five selected OMPs have great potential for vaccine development against S. flexneri infection. The predicted immunogenic epitopes can also be used for development of peptide vaccines or multi-epitope vaccines against human shigellosis. Reverse vaccinology is a promising strategy for the discovery of potential vaccine candidates which can be used for future vaccine development against global persistent infections.
MeSH Terms
-
Anti-Bacterial Agents
beta-Lactam Resistance
Computational Biology
Dysentery, Bacillary
Enterobacteriaceae
Epitopes
Epitopes, T-Lymphocyte
Humans
Membrane Proteins*
Membranes*
Shigella flexneri*
Shigella*
T-Lymphocytes
Vaccines
Vaccines, Subunit
Anti-Bacterial Agents
Epitopes
Epitopes, T-Lymphocyte
Membrane Proteins
Vaccines
Vaccines, Subunit
Figure
Reference
-
1. Kotloff KL, Riddle MS, Platts-Mills JA, Pavlinac P, Zaidi AK. Shigellosis. Lancet. 2018; 391:801–812. PMID: 29254859.
Article2. Gentle A, Ashton PM, Dallman TJ, Jenkins C. Evaluation of molecular methods for serotyping Shigella flexneri. J Clin Microbiol. 2016; 54:1456–1461. PMID: 26984974.
Article3. Killackey SA, Sorbara MT, Girardin SE. Cellular aspects of Shigella pathogenesis: focus on the manipulation of host cell processes. Front Cell Infect Microbiol. 2016; 6:38. PMID: 27066460.
Article4. Khan WA, Griffiths JK, Bennish ML. Gastrointestinal and extra-intestinal manifestations of childhood shigellosis in a region where all four species of Shigella are endemic. PLoS One. 2013; 8:e64097. PMID: 23691156.
Article5. Banga Singh KK, Ojha SC, Deris ZZ, Rahman RA. A 9-year study of shigellosis in Northeast Malaysia: antimicrobial susceptibility and shifting species dominance. Z Gesundh Wiss. 2011; 19:231–236. PMID: 21654922.
Article6. Puzari M, Sharma M, Chetia P. Emergence of antibiotic resistant Shigella species: a matter of concern. J Infect Public Health. 2018; 11:451–454. PMID: 29066021.
Article7. Ashkenazi S, Cohen D. An update on vaccines against Shigella. Ther Adv Vaccines. 2013; 1:113–123. PMID: 24757519.
Article8. Harikrishnan H, Banga Singh KK, Ismail A. Outer membrane proteins analysis of Shigella sonnei and evaluation of their antigenicity in Shigella infected individuals. PLoS One. 2017; 12:e0182878. PMID: 28846684.
Article9. Mani S, Wierzba T, Walker RI. Status of vaccine research and development for Shigella. Vaccine. 2016; 34:2887–2894. PMID: 26979135.
Article10. Pore D, Mahata N, Pal A, Chakrabarti MK. Outer membrane protein A (OmpA) of Shigella flexneri 2a, induces protective immune response in a mouse model. PLoS One. 2011; 6:e22663. PMID: 21818362.
Article11. Tettelin H. The bacterial pan-genome and reverse vaccinology. Genome Dyn. 2009; 6:35–47. PMID: 19696492.
Article12. Pizza M, Scarlato V, Masignani V, et al. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science. 2000; 287:1816–1820. PMID: 10710308.
Article13. He Y, Xiang Z, Mobley HL. Vaxign: the first web-based vaccine design program for reverse vaccinology and applications for vaccine development. J Biomed Biotechnol. 2010; 2010:297505. PMID: 20671958.
Article14. Gardy JL, Laird MR, Chen F, et al. PSORTb v.2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics. 2005; 21:617–623. PMID: 15501914.
Article15. Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001; 305:567–580. PMID: 11152613.16. Sachdeva G, Kumar K, Jain P, Ramachandran S. SPAAN: a software program for prediction of adhesins and adhesin-like proteins using neural networks. Bioinformatics. 2005; 21:483–491. PMID: 15374866.
Article17. Doytchinova IA, Flower DR. VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinformatics. 2007; 8:4. PMID: 17207271.
Article18. Naz A, Awan FM, Obaid A, et al. Identification of putative vaccine candidates against Helicobacter pylori exploiting exoproteome and secretome: a reverse vaccinology based approach. Infect Genet Evol. 2015; 32:280–291. PMID: 25818402.
Article19. Nielsen H. Predicting secretory proteins with SignalP. Methods Mol Biol. 2017; 1611:59–73. PMID: 28451972.
Article20. Wilkins MR, Gasteiger E, Bairoch A, et al. Protein identification and analysis tools in the ExPASy server. Methods Mol Biol. 1999; 112:531–552. PMID: 10027275.
Article21. Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015; 43(Database issue):D447–D452. PMID: 25352553.
Article22. El-Manzalawy Y, Dobbs D, Honavar V. Predicting linear B-cell epitopes using string kernels. J Mol Recognit. 2008; 21:243–255. PMID: 18496882.23. Wang X, Sun Q, Ye Z, et al. Computational approach for predicting the conserved B-cell epitopes of hemagglutinin H7 subtype influenza virus. Exp Ther Med. 2016; 12:2439–2446. PMID: 27703505.
Article24. Singh H, Raghava GP. ProPred: prediction of HLA-DR binding sites. Bioinformatics. 2001; 17:1236–1237. PMID: 11751237.
Article25. Singh H, Raghava GP. ProPred1: prediction of promiscuous MHC class-I binding sites. Bioinformatics. 2003; 19:1009–1014. PMID: 12761064.
Article26. Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000; 28:33–36. PMID: 10592175.
Article27. Levine MM, Kotloff KL, Barry EM, Pasetti MF, Sztein MB. Clinical trials of Shigella vaccines: two steps forward and one step back on a long, hard road. Nat Rev Microbiol. 2007; 5:540–553. PMID: 17558427.
Article28. Rappuoli R, Covacci A. Reverse vaccinology and genomics. Science. 2003; 302:602. PMID: 14576423.
Article29. Oprea M, Antohe F. Reverse-vaccinology strategy for designing T-cell epitope candidates for Staphylococcus aureus endocarditis vaccine. Biologicals. 2013; 41:148–153. PMID: 23582120.30. Rollauer SE, Sooreshjani MA, Noinaj N, Buchanan SK. Outer membrane protein biogenesis in Gram-negative bacteria. Philos Trans R Soc Lond B Biol Sci. 2015; 370:20150023. PMID: 26370935.
Article31. Maldonado RF, Sa-Correia I, Valvano MA. Lipopolysaccharide modification in Gram-negative bacteria during chronic infection. FEMS Microbiol Rev. 2016; 40:480–493. PMID: 27075488.
Article32. Jennison AV, Raqib R, Verma NK. Immunoproteome analysis of soluble and membrane proteins of Shigella flexneri 2457T. World J Gastroenterol. 2006; 12:6683–6688. PMID: 17075984.33. Jarzab A, Witkowska D, Ziomek E, et al. Cyclic OmpC peptidic epitope conjugated to tetanus toxoid as a potential vaccine candidate against shigellosis. Vaccine. 2018; 36:4641–4649. PMID: 29960802.
Article34. Mathavan I, Beis K. The role of bacterial membrane proteins in the internalization of microcin MccJ25 and MccB17. Biochem Soc Trans. 2012; 40:1539–1543. PMID: 23176513.
Article35. Livermore DM. beta-Lactamases in laboratory and clinical resistance. Clin Microbiol Rev. 1995; 8:557–584. PMID: 8665470.
Article36. Raja SB, Murali MR, Devaraj SN. Differential expression of ompC and ompF in multidrug-resistant Shigella dysenteriae and Shigella flexneri by aqueous extract of Aegle marmelos, altering its susceptibility toward beta-lactam antibiotics. Diagn Microbiol Infect Dis. 2008; 61:321–328. PMID: 18358664.37. Levy SB. Active efflux mechanisms for antimicrobial resistance. Antimicrob Agents Chemother. 1992; 36:695–703. PMID: 1503431.
Article38. Yang H, Duan G, Zhu J, et al. The AcrAB-TolC pump is involved in multidrug resistance in clinical Shigella flexneri isolates. Microb Drug Resist. 2008; 14:245–249. PMID: 19035770.39. Miethke M, Marahiel MA. Siderophore-based iron acquisition and pathogen control. Microbiol Mol Biol Rev. 2007; 71:413–451. PMID: 17804665.
Article40. Ashida H, Ogawa M, Mimuro H, Kobayashi T, Sanada T, Sasakawa C. Shigella are versatile mucosal pathogens that circumvent the host innate immune system. Curr Opin Immunol. 2011; 23:448–455. PMID: 21763117.
Article41. Bagchi AK, Sinha AK, Adhikari R, Mukherjee J. Adaptive immune responses during Shigella dysenteriae type 1 infection: an in vitro stimulation with 57 kDa major antigenic OMP in the presence of anti-CD3 antibody. Mol Cell Biochem. 2010; 338:1–10. PMID: 19915959.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Infection of Extended-Spectrum β-Lactamase Producing Shigella flexneri in Children Attending a Childcare Center in Korea
- Enhanced Type III Secretion System Expression of Atypical
Shigella flexneri II:(3)4,7(8) - Epidemiological Characterization of Shigella flexneri Isolates in Korea and the Analysis of Pulsed - Field Gel Electrophoresis Patterns
- Shigella flexneri bacteremia: A case report
- New Vaccine Technology for Control of Emerging and Reemerging Infectious Diseases