Elastase-Positive Neutrophils Are Associated With Refractoriness of Chronic Rhinosinusitis With Nasal Polyps in an Asian Population
- Affiliations
-
- 1Department of Otorhinolaryngology-Head and Neck Surgery, Hallym University Chuncheon Sacred Heart Hospital, Chuncheon, Korea.
- 2Institute of New Frontier Research, Hallym University College of Medicine, Chuncheon, Korea.
- 3Department of Otorhinolaryngology, Armed Forces Capital Hospital, Seongnam, Korea.
- 4Department of Otorhinolaryngology-Head and Neck Surgery, Seoul Metropolitan Government-Seoul National University Boramae Medical Center, Seoul National University College of Medicine, Seoul, Korea. kicubi73@gmail.com
- KMID: 2462571
- DOI: http://doi.org/10.4168/aair.2020.12.1.42
Abstract
- PURPOSE
Various immune cells, including eosinophils and neutrophils, are known to contribute to the development of chronic rhinosinusitis with nasal polyps (CRSwNP). However, the current understanding of the role of neutrophils in the development of CRSwNP still remains unclear. Therefore, we investigated risk factors for refractoriness of CRSwNP in an Asian population.
METHODS
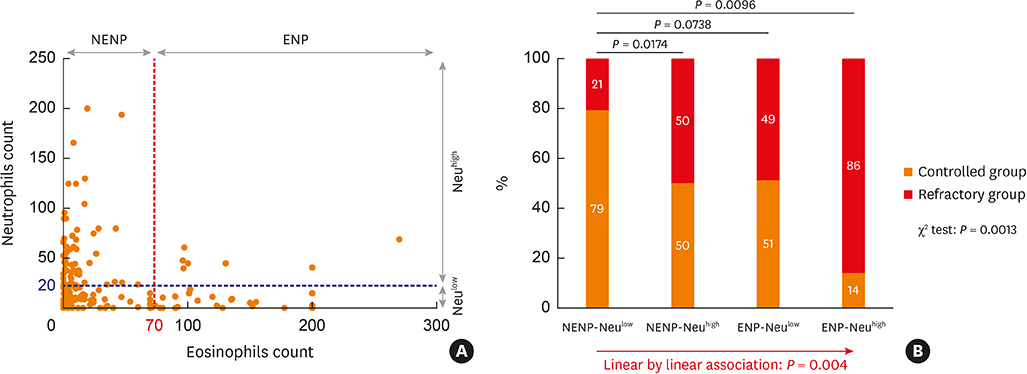
Protein levels of 17 neutrophil-related mediators in nasal polyps (NPs) were determined by multiplex immunoassay, and exploratory factor analysis using principal component analysis was performed. Immunofluorescence analysis was conducted to detect human neutrophil elastase (HNE) or myeloperoxidase (MPO)-positive cells. Tissue eosinophilic nasal polyp (ENP) and tissue neutrophilia (Neu(high)) were defined as greater than 70 eosinophils and 20 HNE-positive cells, otherwise was classified into non-eosinophilic nasal polyp (NENP) and absence of tissue neutrophilia (Neu(low)).
RESULTS
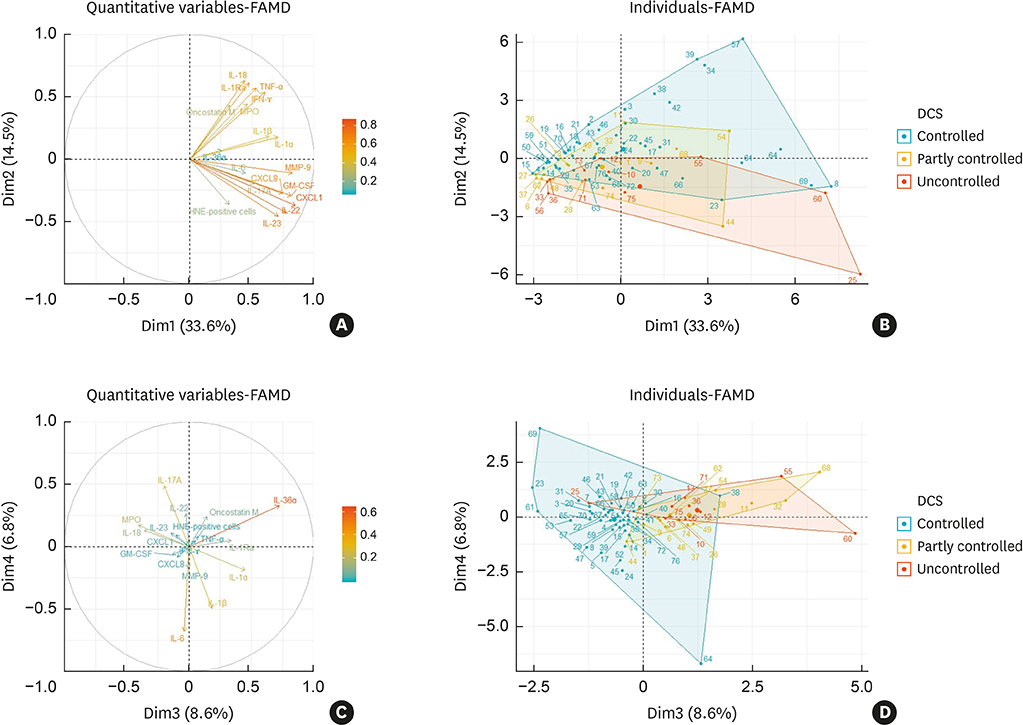
In terms of disease control status, NENP-Neu(low) patients showed the higher rate of disease control than NENP-Neu(high) and ENP-Neu(high) patients. Linear by linear association demonstrated the trend in refractoriness from NENP-Neu(low) to NENP-Neu(high) or ENP-Neu(low) to ENP-Neu(high). When multiple logistic regression was performed, tissue neutrophilia (hazard ratio, 4.38; 95% confidence interval, 1.76-10.85) was found as the strongest risk factor for CRSwNP refractoriness. Additionally, exploratory factor analysis revealed that interleukin (IL)-18, interferon-γ, IL-1Ra, tumor necrosis factor-α, oncostatin M, and MPO were associated with good disease control status, whereas IL-36α and IL-1α were associated with refractory disease control status. In subgroup analysis, HNE-positive cells and IL-36α were significantly upregulated in the refractory group (P = 0.0132 and P = 0.0395, respectively), whereas MPO and IL-18 showed higher expression in the controlled group (P = 0.0002 and P = 0.0009, respectively). Moreover, immunofluorescence analysis revealed that IL-36RâºHNEâº-double positive cells were significantly increased in the refractory group compared to the control group. We also found that the ratio of HNE-positive cells to α1 anti-trypsin was increased in the refractory group.
CONCLUSIONS
Tissue neutrophilia had an influence on treatment outcomes in the Asian CRSwNP patients. HNE-positive cells and IL-36α may be biomarkers for predicting refractoriness in Asians with CRSwNP. Additionally, imbalances in HNE and α1 anti-trypsin may be associated with pathophysiology of neutrophilic chronic rhinosinusitis.
Keyword
MeSH Terms
-
Asian Continental Ancestry Group*
Biomarkers
Eosinophils
Fluorescent Antibody Technique
Humans
Immunoassay
Interleukin 1 Receptor Antagonist Protein
Interleukin-18
Interleukins
Leukocyte Elastase
Logistic Models
Nasal Polyps*
Necrosis
Neutrophils*
Oncostatin M
Peroxidase
Principal Component Analysis
Rhinitis
Risk Factors
Sinusitis
Biomarkers
Interleukin 1 Receptor Antagonist Protein
Interleukin-18
Interleukins
Leukocyte Elastase
Oncostatin M
Peroxidase
Figure
Cited by 5 articles
-
Understanding the Role of Neutrophils in Refractoriness of Chronic Rhinosinusitis With Nasal Polyps
Feng Lan, Luo Zhang
Allergy Asthma Immunol Res. 2020;12(1):1-3. doi: 10.4168/aair.2020.12.1.1.Chinese Society of Allergy and Chinese Society of Otorhinolaryngology-Head and Neck Surgery Guideline for Chronic Rhinosinusitis
Zheng Liu, Jianjun Chen, Lei Cheng, Huabin Li, Shixi Liu, Hongfei Lou, Jianbo Shi, Ying Sun, Dehui Wang, Chengshuo Wang, Xiangdong Wang, Yongxiang Wei, Weiping Wen, Pingchang Yang, Qintai Yang, Gehua Zhang, Yuan Zhang, Changqing Zhao, Dongdong Zhu, Li Zhu, Fenghong Chen, Yi Dong, Qingling Fu, Jingyun Li, Yanqing Li, Chengyao Liu, Feng Liu, Meiping Lu, Yifan Meng, Jichao Sha, Wenyu She, Lili Shi, Kuiji Wang, Jinmei Xue, Luoying Yang, Min Yin, Lichuan Zhang, Ming Zheng, Bing Zhou, Luo Zhang
Allergy Asthma Immunol Res. 2020;12(2):176-237. doi: 10.4168/aair.2020.12.2.176.Clinical Characteristics of Chronic Rhinosinusitis With Nasal Polyp According to Histopathological Endotypes and Staining Method for Neutrophilic Polyp Classification and Its Clinical Implication
Hyoyeon Kim, Shin Hyuk Yoo, Kwang Hyun Byun, Ji-Hun Mo
Korean J Otorhinolaryngol-Head Neck Surg. 2024;67(2):79-86. doi: 10.3342/kjorl-hns.2023.00332.The Correlation of Tissue’s Endotype Biomarkers and Dominant Inflammation Cell in Chronic Rhinosinusitis
Iriana Maharani, Monica Intan, Dyah Indrasworo, Kenty Wantri Anita
Korean J Otorhinolaryngol-Head Neck Surg. 2024;67(3):152-158. doi: 10.3342/kjorl-hns.2023.00493.Analysis of Clinical Characteristics and Post-Treatment Prognosis of Eosinophilic and Neutrophilic Chronic Rhinosinusitis With Nasal Polyps in Korean Patients
Chae-Young Kim, Kwang-Hyun Byun, Jun-Sang Bae, Eun-Hee Kim, Shin-Hyuk Yoo, Ji-Hun Mo
Korean J Otorhinolaryngol-Head Neck Surg. 2024;67(6):328-335. doi: 10.3342/kjorl-hns.2023.00857.
Reference
-
1. Fokkens WJ, Lund VJ, Mullol J, Bachert C, Alobid I, Baroody F, et al. EPOS 2012: European position paper on rhinosinusitis and nasal polyps 2012. A summary for otorhinolaryngologists. Rhinology. 2012; 50:1–12.
Article2. Chen Y, Dales R, Lin M. The epidemiology of chronic rhinosinusitis in Canadians. Laryngoscope. 2003; 113:1199–1205.
Article3. Blackwell DL, Lucas JW, Clarke TC. Summary health statistics for U.S. adults: national health interview survey, 2012. Vital Health Stat 10. 2014; 1–161.4. Hastan D, Fokkens WJ, Bachert C, Newson RB, Bislimovska J, Bockelbrink A, et al. Chronic rhinosinusitis in Europe--an underestimated disease. A GA2LEN study. Allergy. 2011; 66:1216–1223.5. Kim YS, Kim NH, Seong SY, Kim KR, Lee GB, Kim KS. Prevalence and risk factors of chronic rhinosinusitis in Korea. Am J Rhinol Allergy. 2011; 25:117–121.
Article6. Caulley L, Thavorn K, Rudmik L, Cameron C, Kilty SJ. Direct costs of adult chronic rhinosinusitis by using 4 methods of estimation: Results of the US Medical Expenditure Panel Survey. J Allergy Clin Immunol. 2015; 136:1517–1522.
Article7. Halawi AM, Smith SS, Chandra RK. Chronic rhinosinusitis: epidemiology and cost. Allergy Asthma Proc. 2013; 34:328–334.
Article8. Smith KA, Orlandi RR, Rudmik L. Cost of adult chronic rhinosinusitis: a systematic review. Laryngoscope. 2015; 125:1547–1556.
Article9. Van Zele T, Claeys S, Gevaert P, Van Maele G, Holtappels G, Van Cauwenberge P, et al. Differentiation of chronic sinus diseases by measurement of inflammatory mediators. Allergy. 2006; 61:1280–1289.
Article10. Polzehl D, Moeller P, Riechelmann H, Perner S. Distinct features of chronic rhinosinusitis with and without nasal polyps. Allergy. 2006; 61:1275–1279.
Article11. Van Bruaene N, Pérez-Novo CA, Basinski TM, Van Zele T, Holtappels G, De Ruyck N, et al. T-cell regulation in chronic paranasal sinus disease. J Allergy Clin Immunol. 2008; 121:1435–1441. 1441.e1–1433.
Article12. Cao PP, Li HB, Wang BF, Wang SB, You XJ, Cui YH, et al. Distinct immunopathologic characteristics of various types of chronic rhinosinusitis in adult Chinese. J Allergy Clin Immunol. 2009; 124:478–484. 484.e1–472.
Article13. Shi LL, Xiong P, Zhang L, Cao PP, Liao B, Lu X, et al. Features of airway remodeling in different types of Chinese chronic rhinosinusitis are associated with inflammation patterns. Allergy. 2013; 68:101–109.
Article14. Kim DK, Eun KM, Kim MK, Cho D, Han SA, Han SY, et al. Comparison between signature cytokines of nasal tissues in subtypes of chronic rhinosinusitis. Allergy Asthma Immunol Res. 2019; 11:201–211.
Article15. Pothoven KL, Norton JE, Suh LA, Carter RG, Harris KE, Biyasheva A, et al. Neutrophils are a major source of the epithelial barrier disrupting cytokine oncostatin M in patients with mucosal airways disease. J Allergy Clin Immunol. 2017; 139:1966–1978.e9.
Article16. Wen W, Liu W, Zhang L, Bai J, Fan Y, Xia W, et al. Increased neutrophilia in nasal polyps reduces the response to oral corticosteroid therapy. J Allergy Clin Immunol. 2012; 129:1522–1528.e5.
Article17. Wang H, Li ZY, Jiang WX, Liao B, Zhai GT, Wang N, et al. The activation and function of IL-36γ in neutrophilic inflammation in chronic rhinosinusitis. J Allergy Clin Immunol. 2018; 141:1646–1658.
Article18. Georgalas C, Videler W, Freling N, Fokkens W. Global Osteitis Scoring Scale and chronic rhinosinusitis: a marker of revision surgery. Clin Otolaryngol. 2010; 35:455–461.
Article19. Kim DW, Kim JY, Jeon SY. The status of the olfactory cleft may predict postoperative olfactory function in chronic rhinosinusitis with nasal polyposis. Am J Rhinol Allergy. 2011; 25:e90–e94.
Article20. Tokunaga T, Sakashita M, Haruna T, Asaka D, Takeno S, Ikeda H, et al. Novel scoring system and algorithm for classifying chronic rhinosinusitis: the JESREC Study. Allergy. 2015; 70:995–1003.21. Ikeda K, Shiozawa A, Ono N, Kusunoki T, Hirotsu M, Homma H, et al. Subclassification of chronic rhinosinusitis with nasal polyp based on eosinophil and neutrophil. Laryngoscope. 2013; 123:E1–9.
Article22. Kim DK, Jin HR, Eun KM, Mo JH, Cho SH, Oh S, et al. The role of interleukin-33 in chronic rhinosinusitis. Thorax. 2017; 72:635–645.
Article23. Kim DK, Jin HR, Eun KM, Mutusamy S, Cho SH, Oh S, et al. Non-eosinophilic nasal polyps shows increased epithelial proliferation and localized disease pattern in the early stage. PLoS One. 2015; 10:e0139945.
Article24. Kim DW, Kulka M, Jo A, Eun KM, Arizmendi N, Tancowny BP, et al. Cross-talk between human mast cells and epithelial cells by IgE-mediated periostin production in eosinophilic nasal polyps. J Allergy Clin Immunol. 2017; 139:1692–1695.e6.
Article25. Kim DW, Eun KM, Roh EY, Shin S, Kim DK. Chronic rhinosinusitis without nasal polyps in Asian patients shows mixed inflammatory patterns and neutrophil-related disease severity. Mediators Inflamm. 2019; 2019:7138643.
Article26. Van Crombruggen K, Zhang N, Gevaert P, Tomassen P, Bachert C. Pathogenesis of chronic rhinosinusitis: inflammation. J Allergy Clin Immunol. 2011; 128:728–732.
Article27. Gevaert E, Zhang N, Krysko O, Lan F, Holtappels G, De Ruyck N, et al. Extracellular eosinophilic traps in association with Staphylococcus aureus at the site of epithelial barrier defects in patients with severe airway inflammation. J Allergy Clin Immunol. 2017; 139:1849–1860.e6.28. Schleimer RP. Immunopathogenesis of chronic rhinosinusitis and nasal polyposis. Annu Rev Pathol. 2017; 12:331–357.
Article29. Stevens WW, Ocampo CJ, Berdnikovs S, Sakashita M, Mahdavinia M, Suh L, et al. Cytokines in chronic rhinosinusitis. Role in eosinophilia and aspirin-exacerbated respiratory disease. Am J Respir Crit Care Med. 2015; 192:682–694.
Article30. Janciauskiene S, Wrenger S, Immenschuh S, Olejnicka B, Greulich T, Welte T, et al. The multifaceted effects of alpha1-antitrypsin on neutrophil functions. Front Pharmacol. 2018; 9:341.
Article31. Meijer M, Rijkers GT, van Overveld FJ. Neutrophils and emerging targets for treatment in chronic obstructive pulmonary disease. Expert Rev Clin Immunol. 2013; 9:1055–1068.
Article32. Lou H, Meng Y, Piao Y, Wang C, Zhang L, Bachert C. Predictive significance of tissue eosinophilia for nasal polyp recurrence in the Chinese population. Am J Rhinol Allergy. 2015; 29:350–356.
Article33. Cao Y, Chen F, Sun Y, Hong H, Wen Y, Lai Y, et al. LL-37 promotes neutrophil extracellular trap formation in chronic rhinosinusitis with nasal polyps. Clin Exp Allergy. 2019; 49:990–999.
Article34. Tomassen P, Vandeplas G, Van Zele T, Cardell LO, Arebro J, Olze H, et al. Inflammatory endotypes of chronic rhinosinusitis based on cluster analysis of biomarkers. J Allergy Clin Immunol. 2016; 137:1449–1456.e4.
Article35. Liao B, Liu JX, Li ZY, Zhen Z, Cao PP, Yao Y, et al. Multidimensional endotypes of chronic rhinosinusitis and their association with treatment outcomes. Allergy. 2018; 73:1459–1469.
Article36. Mayadas TN, Cullere X, Lowell CA. The multifaceted functions of neutrophils. Annu Rev Pathol. 2014; 9:181–218.
Article37. Soehnlein O, Steffens S, Hidalgo A, Weber C. Neutrophils as protagonists and targets in chronic inflammation. Nat Rev Immunol. 2017; 17:248–261.
Article38. Van Zele T, Holtappels G, Gevaert P, Bachert C. Differences in initial immunoprofiles between recurrent and nonrecurrent chronic rhinosinusitis with nasal polyps. Am J Rhinol Allergy. 2014; 28:192–198.
Article39. Faurschou M, Borregaard N. Neutrophil granules and secretory vesicles in inflammation. Microbes Infect. 2003; 5:1317–1327.
Article40. Campanelli D, Melchior M, Fu Y, Nakata M, Shuman H, Nathan C, et al. Cloning of cDNA for proteinase 3: a serine protease, antibiotic, and autoantigen from human neutrophils. J Exp Med. 1990; 172:1709–1715.
Article41. Witko-Sarsat V, Rieu P, Descamps-Latscha B, Lesavre P, Halbwachs-Mecarelli L. Neutrophils: molecules, functions and pathophysiological aspects. Lab Invest. 2000; 80:617–653.
Article42. Janciauskiene SM, Bals R, Koczulla R, Vogelmeier C, Köhnlein T, Welte T. The discovery of α1-antitrypsin and its role in health and disease. Respir Med. 2011; 105:1129–1139.
Article43. Kim S, Nadel JA. Role of neutrophils in mucus hypersecretion in COPD and implications for therapy. Treat Respir Med. 2004; 3:147–159.
Article44. Vlahos R, Wark PA, Anderson GP, Bozinovski S. Glucocorticosteroids differentially regulate MMP-9 and neutrophil elastase in COPD. PLoS One. 2012; 7:e33277.
Article45. Ryu G, Kim DK, Dhong HJ, Eun KM, Lee KE, Kong IG, et al. Immunological characteristics in refractory chronic rhinosinusitis with nasal polyps undergoing revision surgeries. Allergy Asthma Immunol Res. 2019; 11:664–676.
Article46. Kim DK, Kang SI, Kong IG, Cho YH, Song SK, Hyun SJ, et al. Two-track medical treatment strategy according to the clinical scoring system for chronic rhinosinusitis. Allergy Asthma Immunol Res. 2018; 10:490–502.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Understanding the Role of Neutrophils in Refractoriness of Chronic Rhinosinusitis With Nasal Polyps
- Medical treatment according to phenotypes of chronic rhinosinusitis
- Biomarkers in Chronic Rhinosinusitis with Nasal Polyp: Personalized Medicine Based on Endotype
- Neutrophils as a Protagonist and Target in Chronic Rhinosinusitis
- Clinical Characteristics of Chronic Rhinosinusitis With Nasal Polyp According to Histopathological Endotypes and Staining Method for Neutrophilic Polyp Classification and Its Clinical Implication