Endocrinol Metab.
2019 Sep;34(3):314-322. 10.3803/EnM.2019.34.3.314.
Transcriptome Analysis Identifies an Attenuated Local Immune Response in Invasive Nonfunctioning Pituitary Adenomas
- Affiliations
-
- 1Department of Neurosurgery, Seoul National University Hospital, Seoul National University College of Medicine, Seoul, Korea.
- 2Pituitary Center, Seoul National University Hospital, Seoul National University College of Medicine, Seoul, Korea. jhkxingfu@gmail.com
- 3Department of Internal Medicine, Seoul National University Hospital, Seoul National University College of Medicine, Seoul, Korea.
- 4Department of Molecular Medicine and Biopharmaceutical Sciences, Graduate School of Convergence Science and Technology, Seoul National University College of Medicine, Seoul, Korea.
- KMID: 2458634
- DOI: http://doi.org/10.3803/EnM.2019.34.3.314
Abstract
- BACKGROUND
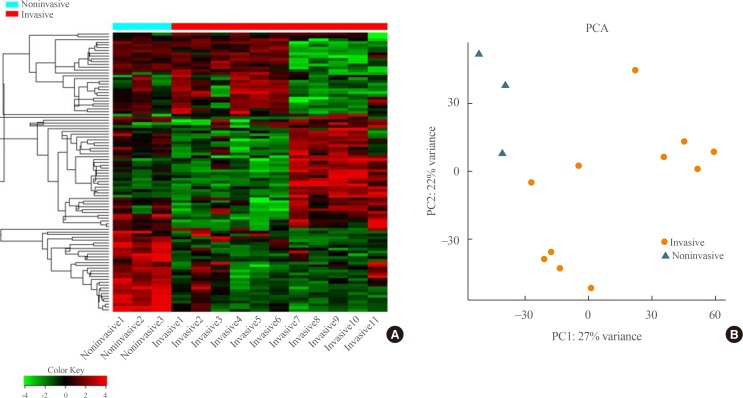
Invasive nonfunctioning pituitary adenomas (NFPAs) remain challenging due to their high complication rate and poor prognosis. We aimed to identify the distinctive molecular signatures of invasive NFPAs, compared with noninvasive NFPAs, using gene expression profiling by RNA sequencing.
METHODS
We obtained frozen fresh tissue samples from 14 patients with NFPAs who underwent primary transsphenoidal surgery. Three non-invasive and 11 invasive NFPAs were used for RNA sequencing. The bioinformatics analysis included differential gene expression, gene ontology analysis, and pathway analysis.
RESULTS
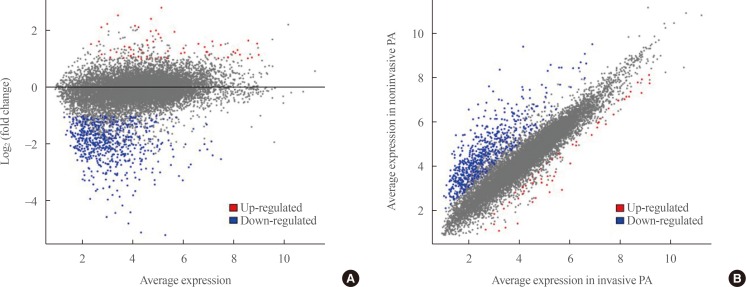
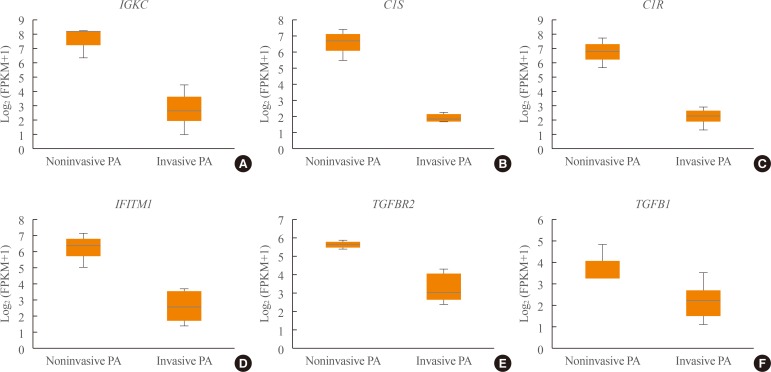
A total of 700 genes were differentially expressed (59 up-regulated and 641 down-regulated genes) between invasive and non-invasive NFPAs (false discovery rate <0.1, and |fold change|≥2). Using the down-regulated genes in invasive NFPAs, gene ontology enrichment analyses and pathway analyses demonstrated that the local immune response was attenuated and that transforming growth factor-β (TGF-β) RII-initiated TGF-β signaling was down-regulated in invasive NFPAs. The overexpression of claudin-9 (CLDN9) and the down-regulation of insulin-like growth factor-binding protein 5 (IGFBP5), death-associated protein kinase 1 (DAPK1), and tissue inhibitor of metalloproteinase-3 (TIMP3) may be related with invasiveness in NFPAs.
CONCLUSION
Invasive NFPAs harbor different gene expression profiles relative to noninvasive NFPAs. In particular, local suppression of the immune response and TGF-β signaling can make PAs prone to invasiveness.
MeSH Terms
Figure
Reference
-
1. Hansen TM, Batra S, Lim M, Gallia GL, Burger PC, Salvatori R, et al. Invasive adenoma and pituitary carcinoma: a SEER database analysis. Neurosurg Rev. 2014; 37:279–285. PMID: 24526366.
Article2. Kim JH, Lee JH, Lee JH, Hong AR, Kim YJ, Kim YH. Endoscopic transsphenoidal surgery outcomes in 331 nonfunctioning pituitary adenoma cases after a single surgeon learning curve. World Neurosurg. 2018; 109:e409–e416. PMID: 29017983.
Article3. Trouillas J, Roy P, Sturm N, Dantony E, Cortet-Rudelli C, Viennet G, et al. A new prognostic clinicopathological classification of pituitary adenomas: a multicentric case-control study of 410 patients with 8 years post-operative follow-up. Acta Neuropathol. 2013; 126:123–135. PMID: 23400299.4. Mete O, Ezzat S, Asa SL. Biomarkers of aggressive pituitary adenomas. J Mol Endocrinol. 2012; 49:R69–R78. PMID: 22822048.
Article5. Yang Q, Li X. Molecular network basis of invasive pituitary adenoma: a review. Front Endocrinol (Lausanne). 2019; 10:7. PMID: 30733705.
Article6. Knosp E, Steiner E, Kitz K, Matula C. Pituitary adenomas with invasion of the cavernous sinus space: a magnetic resonance imaging classification compared with surgical findings. Neurosurgery. 1993; 33:610–617. PMID: 8232800.7. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. PMID: 23104886.
Article8. Anders S, Pyl PT, Huber W. HTSeq: a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015; 31:166–169. PMID: 25260700.9. Ge SX, Son EW, Yao R. iDEP: an integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinformatics. 2018; 19:534. PMID: 30567491.
Article10. Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016; 44:W90–W97. PMID: 27141961.
Article11. Richardson TE, Shen ZJ, Kanchwala M, Xing C, Filatenkov A, Shang P, et al. Aggressive behavior in silent subtype III pituitary adenomas may depend on suppression of local immune response: a whole transcriptome analysis. J Neuropathol Exp Neurol. 2017; 76:874–882. PMID: 28922848.
Article12. Yang Q, Wang Y, Zhang S, Tang J, Li F, Yin J, et al. Biomarker discovery for immunotherapy of pituitary adenomas: enhanced robustness and prediction ability by modern computational tools. Int J Mol Sci. 2019; 20:E151. PMID: 30609812.
Article13. Mei Y, Bi WL, Greenwald NF, Du Z, Agar NY, Kaiser UB, et al. Increased expression of programmed death ligand 1 (PD-L1) in human pituitary tumors. Oncotarget. 2016; 7:76565–76576. PMID: 27655724.
Article14. Zhenye L, Chuzhong L, Youtu W, Xiaolei L, Lei C, Lichuan H, et al. The expression of TGF-β1, Smad3, phospho-Smad3 and Smad7 is correlated with the development and invasion of nonfunctioning pituitary adenomas. J Transl Med. 2014; 12:71. PMID: 24636138.
Article15. Gu YH, Feng YG. Down-regulation of TGF-β RII expression is correlated with tumor growth and invasion in non-functioning pituitary adenomas. J Clin Neurosci. 2018; 47:264–268. PMID: 29031543.
Article16. Galland F, Lacroix L, Saulnier P, Dessen P, Meduri G, Bernier M, et al. Differential gene expression profiles of invasive and non-invasive non-functioning pituitary adenomas based on microarray analysis. Endocr Relat Cancer. 2010; 17:361–371. PMID: 20228124.
Article17. Hwang JR, Cho YJ, Lee Y, Park Y, Han HD, Ahn HJ, et al. The C-terminus of IGFBP-5 suppresses tumor growth by inhibiting angiogenesis. Sci Rep. 2016; 6:39334. PMID: 28008951.
Article18. de Araujo LJ, Lerario AM, de Castro M, Martins CS, Bronstein MD, Machado MC, et al. Transcriptome analysis showed a differential signature between invasive and non-invasive corticotrophinomas. Front Endocrinol (Lausanne). 2017; 8:55. PMID: 28382019.
Article19. Rosas SL, Koch W, da Costa Carvalho MG, Wu L, Califano J, Westra W, et al. Promoter hypermethylation patterns of p16, O6-methylguanine-DNA-methyltransferase, and death-associated protein kinase in tumors and saliva of head and neck cancer patients. Cancer Res. 2001; 61:939–942. PMID: 11221887.20. Tofrizal A, Fujiwara K, Azuma M, Kikuchi M, Jindatip D, Yashiro T, et al. Tissue inhibitors of metalloproteinase-expressing cells in human anterior pituitary and pituitary adenoma. Med Mol Morphol. 2017; 50:145–154. PMID: 28353090.
Article21. Cao C, Wang W, Ma C, Jiang P. Computational analysis identifies invasion-associated genes in pituitary adenomas. Mol Med Rep. 2015; 12:1977–1982. PMID: 25824863.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Transcriptome Analysis Identifies an Attenuated Local Immune Response in Invasive Nonfunctioning Pituitary Adenomas
- Comparative evaluation of CT and secreting hormones in pituitary adenomas
- Deliberate Two-Staged Endoscopic Endonasal Transsphenoidal Surgery in Large Pituitary Adenomas
- Endoscopic Endonasal Surgery for Pituitary Adenoma
- Pituitary Adenomas in Childhood and Adolescence