Korean J Gastroenterol.
2019 Jul;74(1):30-41. 10.4166/kjg.2019.74.1.30.
Development of a Pancreatic Cancer Specific Binding Peptide Using Phage Display
- Affiliations
-
- 1Catholic Photomedicine Research Institute, Seoul, Korea. parkjerry@catholic.ac.kr
- 2Department of Medical Lifescience, College of Medicine, The Catholic University of Korea, Seoul, Korea.
- 3Department of Internal Medicine, Seoul St. Mary's Hospital, College of Medicine, The Catholic University of Korea, Seoul, Korea.
- KMID: 2453256
- DOI: http://doi.org/10.4166/kjg.2019.74.1.30
Abstract
- BACKGROUND/AIMS
Pancreatic cancer has a very poor prognosis, and early diagnosis is a way to increase the survival rate of patients. The purpose of this study was to develop pancreatic cancer-specific peptides for imaging studies.
METHODS
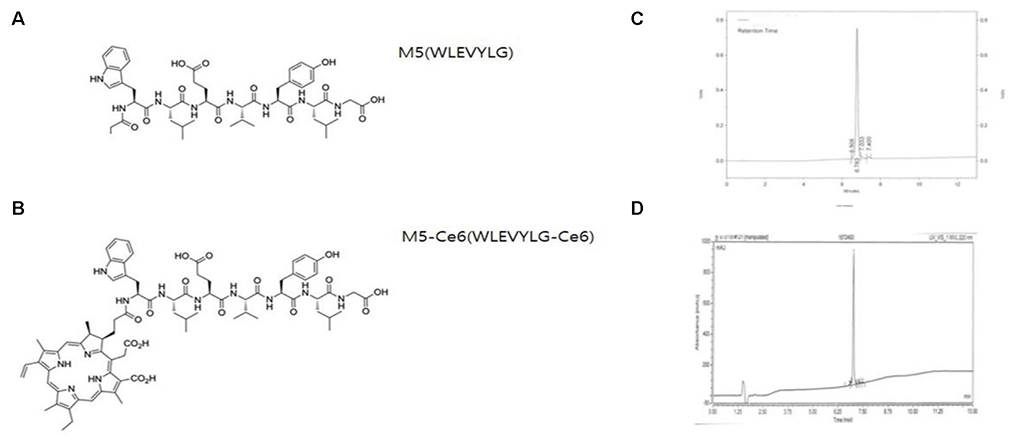
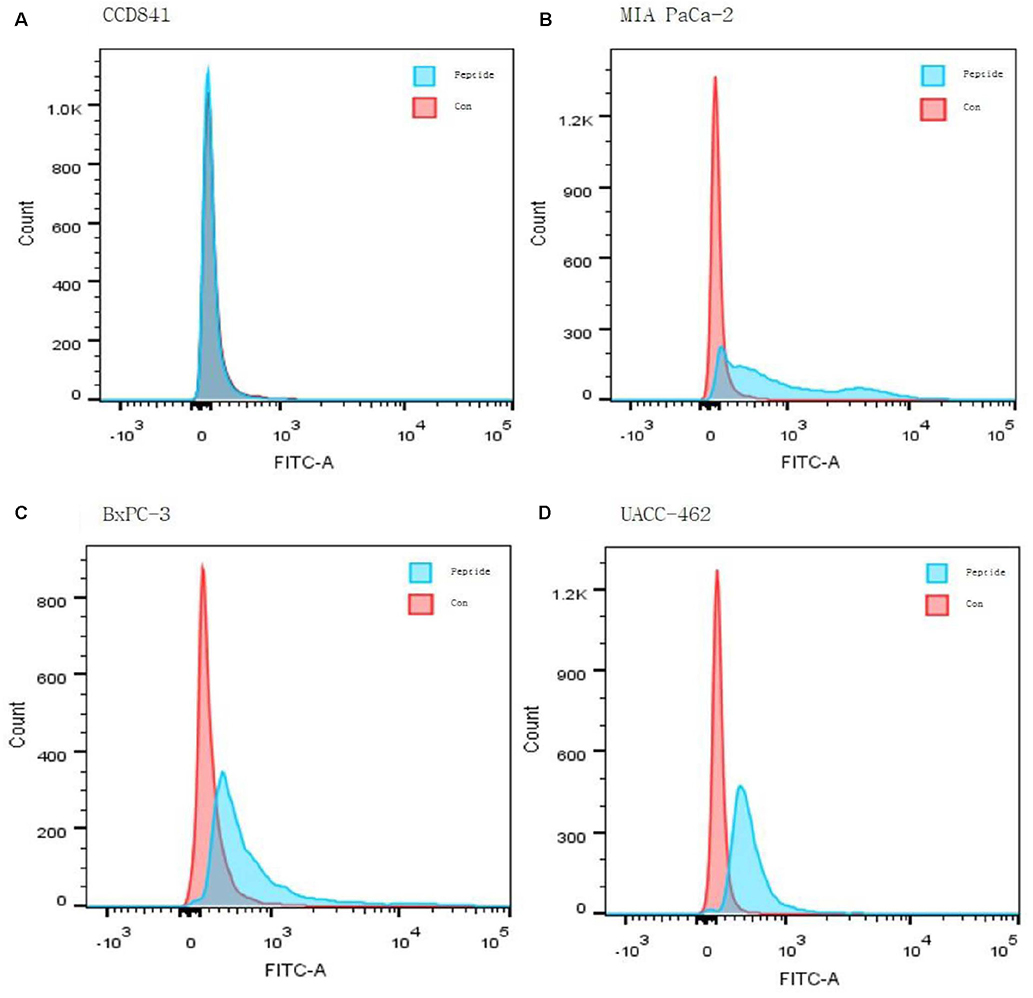
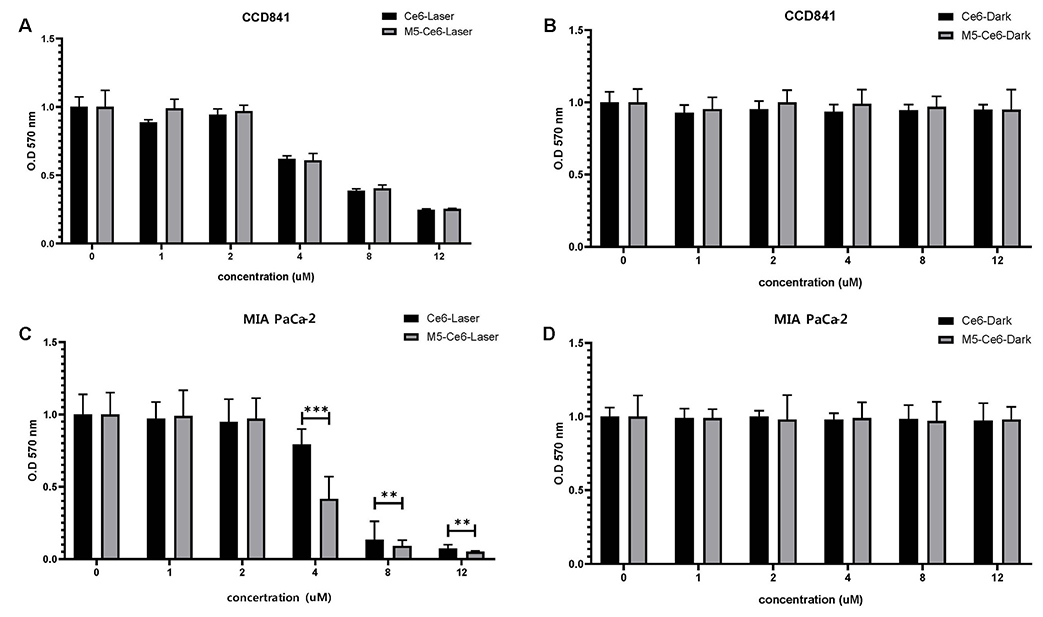
Three pancreatic cancer cell lines, MIA PaCa-2, UACC-462, and BxPC-3, and a control cell line, CCD841, were used. Biopannings were performed on MIA PaCa-2 using a phage display library. After this, the peptides were synthesized and labeled with fluorescein isothiocyanate (FITC). Immunocytochemistry (ICC), enzyme-linked immunosorbent assay (ELISA), and fluorescence-activated cell sorter (FACS) were performed to examine the specific binding. To examine its therapeutic applications, a photosensitizer, chlorin e6 (Ce6), was conjugated on the peptide and photodynamic therapy was performed. Cell survival was investigated using a [3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide] assay.
RESULTS
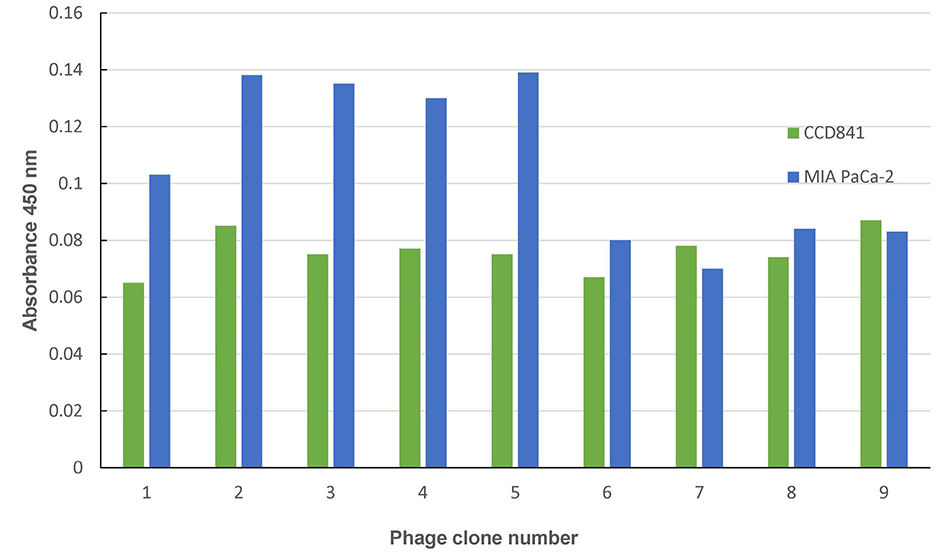
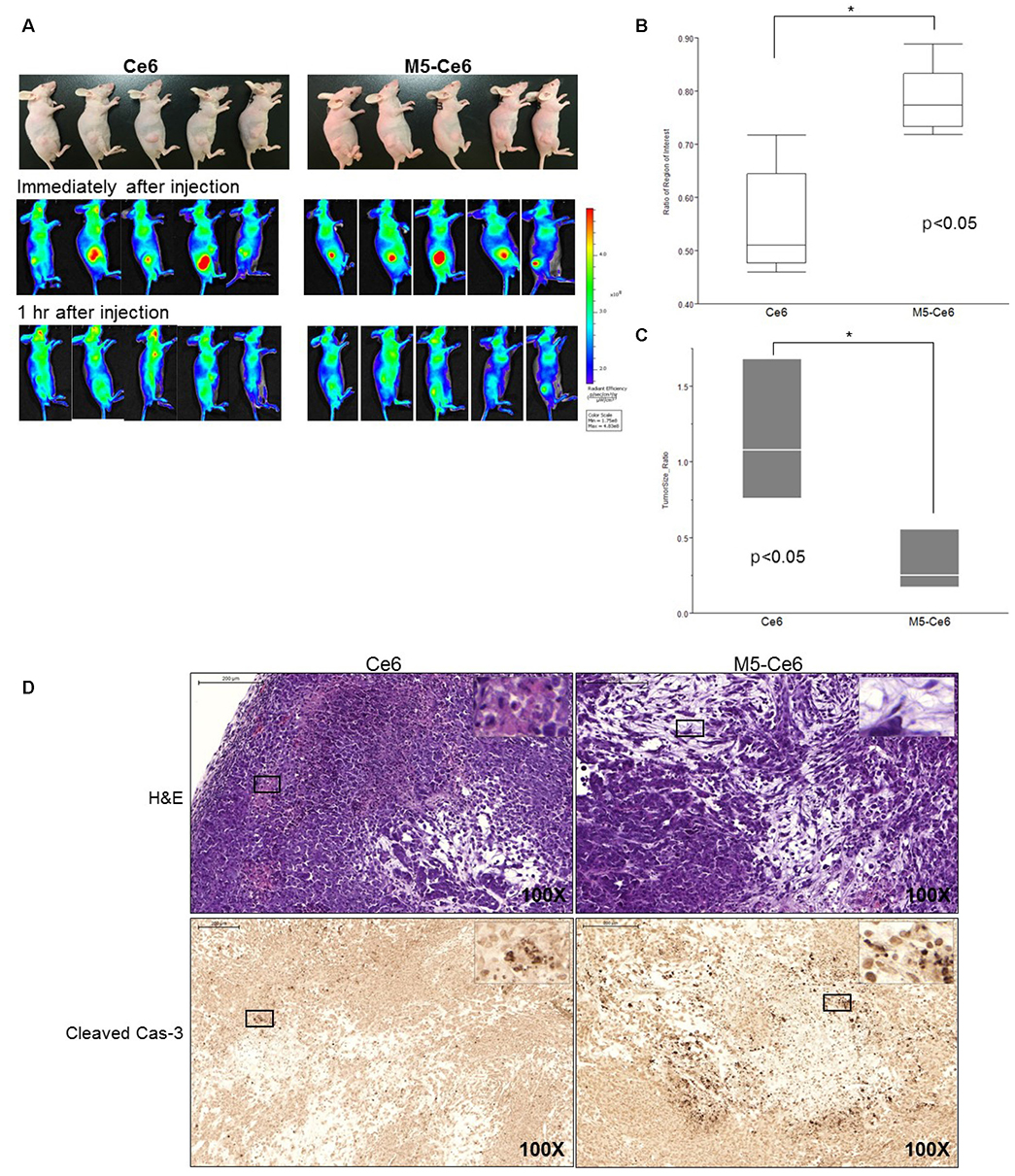
After three biopannings, the phages were amplified from 1.4×104 to 3.2×105 plaque-forming units. The most strongly binding phage was selected from the ELISA and ICC results. FITC-labeled peptide, M5, in the three pancreatic cancer cell lines showed significantly higher immunofluorescence in the ICC experiments than that of CCD841. The higher binding ability to MIA PaCa-2 cells was confirmed from FACS analysis, which showed a right shift compared to CCD841. M5 bound to Ce6 showed a significantly lower cell survival rate than that of Ce6 alone in photodynamic therapy, which was observed consistently as a change in the tumor size and fluorescence intensity in MIA PaCa-2 cell-implanted animal models.
CONCLUSIONS
This study showed that the noble peptide, M5, binds specifically to the pancreatic cancer cell line, MIA PaCa-2. The M5 peptide has potential use in future optical diagnostic and therapeutic purposes.
MeSH Terms
Figure
Reference
-
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018; 68:7–30.
Article2. Ryu JK. The early detection of pancreatic cancer: whom and how? Korean J Pancreas Biliary Tract. 2015; 20:198–203.
Article3. Kamisawa T, Wood LD, Itoi T, Takaori K. Pancreatic cancer. Lancet. 2016; 388:73–85.
Article4. Fong ZV, Tan WP, Lavu H, et al. Preoperative imaging for resectable periampullary cancer: clinicopathologic implications of reported radiographic findings. J Gastrointest Surg. 2013; 17:1098–1106.
Article5. Pannala R, Basu A, Petersen GM, Chari ST. New-onset diabetes: a potential clue to the early diagnosis of pancreatic cancer. Lancet Oncol. 2009; 10:88–95.
Article6. Kozuka S, Sassa R, Taki T, et al. Relation of pancreatic duct hyperplasia to carcinoma. Cancer. 1979; 43:1418–1428.
Article7. Lee JH, Cassani LS, Bhosale P, Ross WA. The endoscopist's role in the diagnosis and management of pancreatic cancer. Expert Rev Gastroenterol Hepatol. 2016; 10:1027–1039.
Article8. Chu LC, Goggins MG, Fishman EK. Diagnosis and detection of pancreatic cancer. Cancer J. 2017; 23:333–342.
Article9. Goonetilleke KS, Siriwardena AK. Systematic review of carbohydrate antigen (CA 19-9) as a biochemical marker in the diagnosis of pancreatic cancer. Eur J Surg Oncol. 2007; 33:266–270.
Article10. Bausch D, Thomas S, Mino-Kenudson M, et al. Plectin-1 as a novel biomarker for pancreatic cancer. Clin Cancer Res. 2011; 17:302–309.
Article11. Faca VM, Song KS, Wang H, et al. A mouse to human search for plasma proteome changes associated with pancreatic tumor development. PLoS Med. 2008; 5:e123.
Article12. Koopmann J, Fedarko NS, Jain A, et al. Evaluation of osteopontin as biomarker for pancreatic adenocarcinoma. Cancer Epidemiol Biomarkers Prev. 2004; 13:487–491.
Article13. Sidhu SS. Phage display in pharmaceutical biotechnology. Curr Opin Biotechnol. 2000; 11:610–616.
Article14. Wilson DR, Finlay BB. Phage display: applications, innovations, and issues in phage and host biology. Can J Microbiol. 1998; 44:313–329.
Article15. Rodi DJ, Makowski L. Phage-display technology--finding a needle in a vast molecular haystack. Curr Opin Biotechnol. 1999; 10:87–93.16. Frei JC, Lai JR. Protein and antibody engineering by phage display. Methods Enzymol. 2016; 580:45–87.
Article17. Joshi BP, Wang TD. Targeted optical imaging agents in cancer: focus on clinical applications. Contrast Media Mol Imaging. 2018; 2018:2015237.
Article18. Kaminski MF, Regula J, Kraszewska E, et al. Quality indicators for colonoscopy and the risk of interval cancer. N Engl J Med. 2010; 362:1795–1803.
Article19. Hsiung PL, Hardy J, Friedland S, et al. Detection of colonic dysplasia in vivo using a targeted heptapeptide and confocal microendoscopy. Nat Med. 2008; 14:454–458.
Article20. Gu YL, Lan C, Pei H, Yang SN, Liu YF, Xiao LL. Applicative value of serum CA19-9, CEA, CA125 and CA242 in diagnosis and prognosis for patients with pancreatic cancer treated by concurrent chemoradiotherapy. Asian Pac J Cancer Prev. 2015; 16:6569–6573.
Article21. Honda K, Katzke VA, Hüsing A, et al. CA19-9 and apolipoprotein-A2 isoforms as detection markers for pancreatic cancer: a prospective evaluation. Int J Cancer. 2019; 144:1877–1887.22. DeWitt J, Devereaux B, Chriswell M, et al. Comparison of endoscopic ultrasonography and multidetector computed tomography for detecting and staging pancreatic cancer. Ann Intern Med. 2004; 141:753–763.
Article23. Joshi BP, Dai Z, Gao Z, et al. Detection of sessile serrated adenomas in the proximal colon using wide-field fluorescence endoscopy. Gastroenterology. 2017; 152:1002–1013.e9.
Article24. Bang JY, Magee SH, Ramesh J, Trevino JM, Varadarajulu S. Randomized trial comparing fanning with standard technique for endoscopic ultrasound-guided fine-needle aspiration of solid pancreatic mass lesions. Endoscopy. 2013; 45:445–450.
Article25. Bang S. Surveillance and early diagnosis of pancreatic cancer. Korean J Pancreas Biliary Tract. 2015; 20:1–4.
Article26. Takaishi S, Okumura T, Tu S, et al. Identification of gastric cancer stem cells using the cell surface marker CD44. Stem Cells. 2009; 27:1006–1020.
Article27. Moris M, Dawson DW, Jiang J, et al. Plectin-1 as a biomarker of malignant progression in intraductal papillary mucinous neoplasms: a multicenter study. Pancreas. 2016; 45:1353–1358.28. Wang J, Zheng M, Xie Z. Carrier-free core-shell nanodrugs for synergistic two-photon photodynamic therapy of cervical cancer. J Colloid Interface Sci. 2019; 535:84–91.
Article29. Zullo SW, Zeitouni NC, Segal RJ. Long-term efficacy of combination vismodegib and photodynamic therapy for multiple basal cell carcinomas. Photodiagnosis Photodyn Ther. 2018; 24:164–165.
Article30. Lee KH. Chemotherapy and targeted therapy with management of related complications in pancreatic cancer. Korean J Pancreas Biliary Tract. 2015; 20:5–13.
Article31. Moore MJ, Goldstein D, Hamm J, et al. Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: a phase III trial of the National Cancer Institute of Canada Clinical Trials Group. J Clin Oncol. 2007; 25:1960–1966.
Article32. Neoptolemos JP, Stocken DD, Friess H, et al. A randomized trial of chemoradiotherapy and chemotherapy after resection of pancreatic cancer. N Engl J Med. 2004; 350:1200–1210.
Article33. Yabar CS, Winter JM. Pancreatic cancer: a review. Gastroenterol Clin North Am. 2016; 45:429–445.34. Huggett MT, Jermyn M, Gillams A, et al. Phase I/II study of verteporfin photodynamic therapy in locally advanced pancreatic cancer. Br J Cancer. 2014; 110:1698–1704.
Article35. Bown SG, Rogowska AZ, Whitelaw DE, et al. Photodynamic therapy for cancer of the pancreas. Gut. 2002; 50:549–557.
Article36. Choi JH, Oh D, Lee JH, et al. Initial human experience of endoscopic ultrasound-guided photodynamic therapy with a novel photosensitizer and a flexible laser-light catheter. Endoscopy. 2015; 47:1035–1038.
Article37. Yang Y, Hou W, Liu S, Sun K, Li M, Wu C. Biodegradable polymer nanoparticles for photodynamic therapy by bioluminescence resonance energy transfer. Biomacromolecules. 2018; 19:201–208.
Article38. Kim YR, Kim S, Choi JW, et al. Bioluminescence-activated deep-tissue photodynamic therapy of cancer. Theranostics. 2015; 5:805–817.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- A Phage Display-Identified Peptide Selectively Binds to Kidney Injury Molecule-1 (KIM-1) and Detects KIM-1–Overexpressing Tumors In Vivo
- Terminal Protein-specific scFv Production by Phage Display
- Establishement of Antibody Selection by Ribosome Display
- Identification of high-affinity VEGFR3-binding peptides through a phage-displayed random peptide library
- Definition of the peptide mimotope of cellular receptor for hepatitis C virus E2 protein using random peptide library