Int J Stem Cells.
2018 Nov;11(2):187-195. 10.15283/ijsc18057.
MiR-9 Controls Chemotactic Activity of Cord Blood CD34⺠Cells by Repressing CXCR4 Expression
- Affiliations
-
- 1Soonchunhyang Institute of Medi-bio Science (SIMS), Soon Chun Hyang University, Cheonan, Korea. leeman@sch.ac.kr, yunklee@sch.ac.kr, yshwang0428@sch.ac.kr
- 2Department of Obstetrics and Gynecology, Soon Chun Hyang University College of Medicine, Bucheon, Korea.
- 3Department of Obstetrics and Gynecology, Soon Chun Hyang University Cheonan Hospital, Cheonan, Korea.
- 4Microbiology and Immunology, Indiana University School of Medicine, Indianapolis, USA.
- KMID: 2429551
- DOI: http://doi.org/10.15283/ijsc18057
Abstract
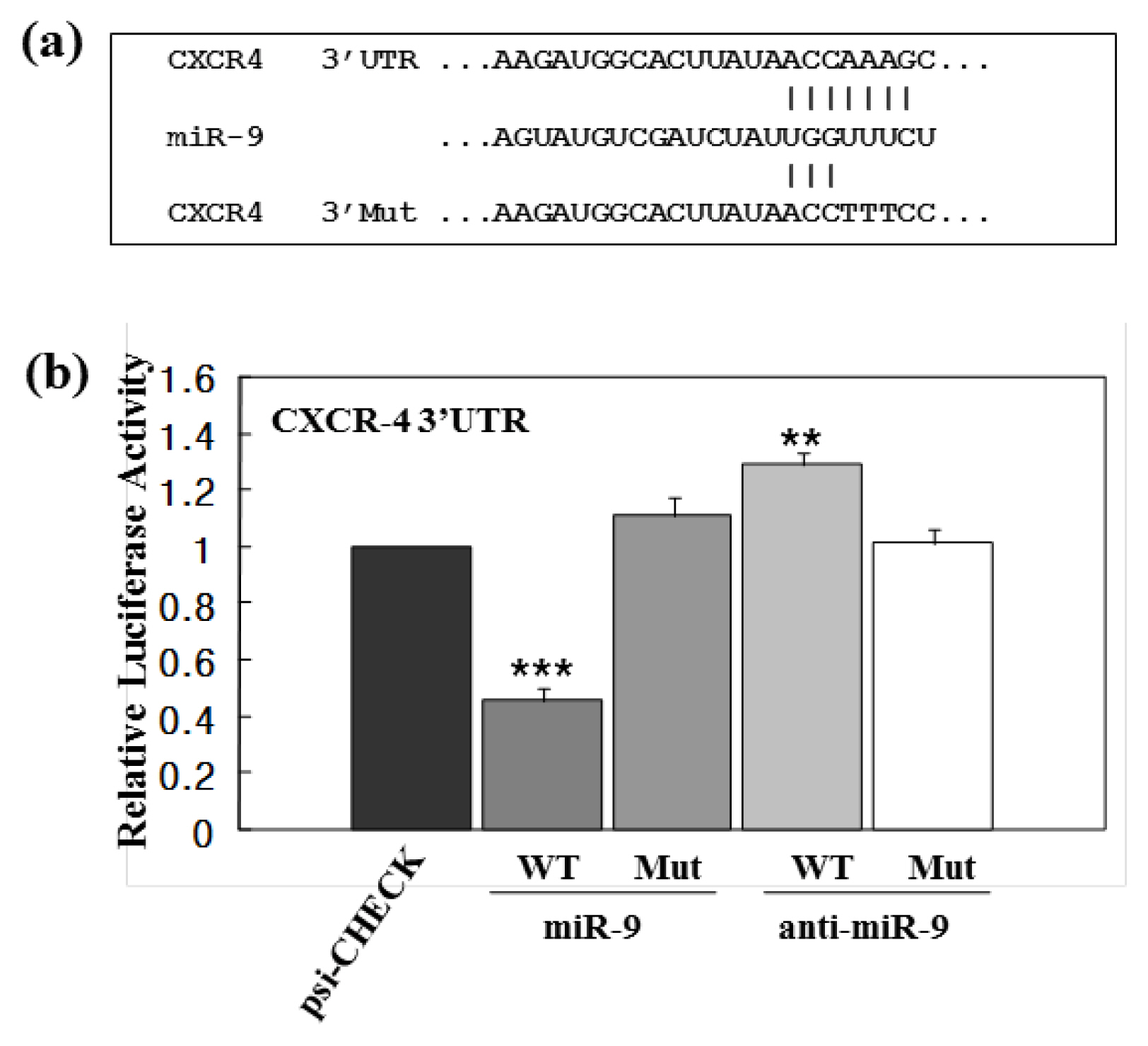
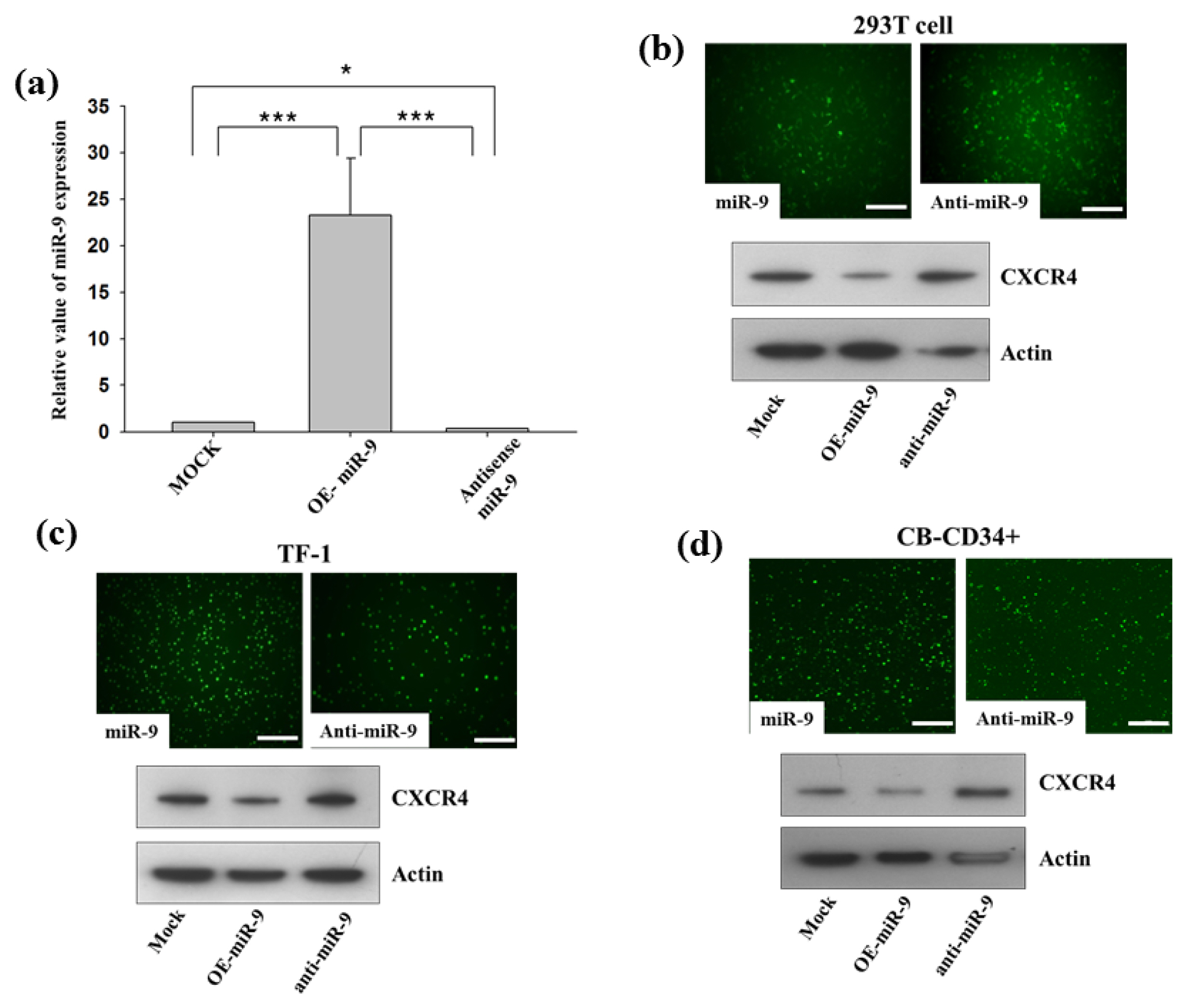
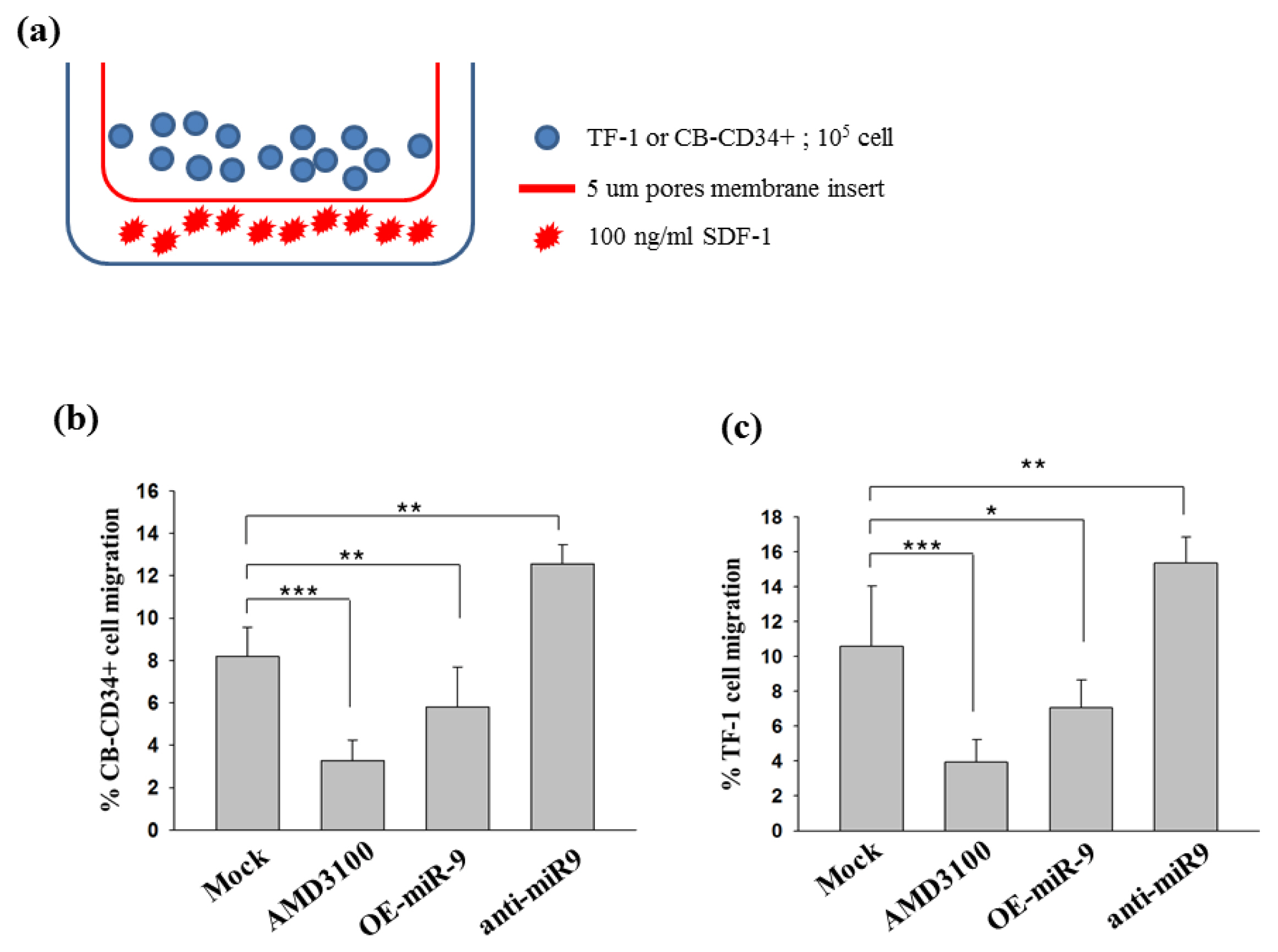
- Improved approaches for promoting umbilical cord blood (CB) hematopoietic stem cell (HSC) homing are clinically important to enhance engraftment of CB-HSCs. Clinical transplantation of CB-HSCs is used to treat a wide range of disorders. However, an improved understanding of HSC chemotaxis is needed for facilitation of the engraftment process. We found that ectopic overexpression of miR-9 and antisense-miR-9 respectively down- and up-regulated C-X-C chemokine receptor type 4 (CXCR4) expression in CB-CD34⺠cells as well as in 293T and TF-1 cell lines. Since CXCR4 is a specific receptor for the stromal cell derived factor-1 (SDF-1) chemotactic factor, we investigated whether sense miR-9 and antisense miR-9 influenced CXCR4-mediated chemotactic mobility of primary CB CD34⺠cells and TF-1 cells. Ectopic overexpression of sense miR-9 and antisense miR-9 respectively down- and up-regulated SDF-1-mediated chemotactic cell mobility. To our knowledge, this study is the first to report that miR-9 may play a role in regulating CXCR4 expression and SDF-1-mediated chemotactic activity of CB CD34⺠cells.
Keyword
MeSH Terms
Figure
Reference
-
References
1. Broxmeyer HE. Chemokines in hematopoiesis. Curr Opin Hematol. 2008; 15:49–58. DOI: 10.1097/MOH.0b013e3282f29012.
Article2. Kollet O, Petit I, Kahn J, Samira S, Dar A, Peled A, Deutsch V, Gunetti M, Piacibello W, Nagler A, Lapidot T. Human CD34(+)CXCR4(-) sorted cells harbor intracellular CXCR4, which can be functionally expressed and provide NOD/SCID repopulation. Blood. 2002; 100:2778–2786. DOI: 10.1182/blood-2002-02-0564. PMID: 12351385.
Article3. Ramirez P, Wagner JE, Defor TE, Eide CR, Miller JS, Weisdorf DJ, Brunstein CG. CXCR4 expression in CD34+ cells and unit predominance after double umbilical cord blood transplantation. Leukemia. 2013; 27:1181–1183. DOI: 10.1038/leu.2012.261. PMCID: 3668782.
Article4. Dar A, Goichberg P, Shinder V, Kalinkovich A, Kollet O, Netzer N, Margalit R, Zsak M, Nagler A, Hardan I, Resnick I, Rot A, Lapidot T. Chemokine receptor CXCR4-dependent internalization and resecretion of functional chemokine SDF-1 by bone marrow endothelial and stromal cells. Nat Immunol. 2006; 6:1038–1046. DOI: 10.1038/ni1251.
Article5. Lapidot T, Dar A, Kollet O. How do stem cells find their way home? Blood. 2005; 106:1901–1910. DOI: 10.1182/blood-2005-04-1417. PMID: 15890683.
Article6. Ma Q, Jones D, Borghesani PR, Segal RA, Nagasawa T, Kishimoto T, Bronson RT, Springer TA. Impaired B-lymphopoiesis, myelopoiesis, and derailed cerebellar neuron migration in CXCR4- and SDF-1-deficient mice. Proc Natl Acad Sci U S A. 1998; 95:9448–9453. DOI: 10.1073/pnas.95.16.9448. PMID: 9689100. PMCID: 21358.
Article7. Ponomaryov T, Peled A, Petit I, Taichman RS, Habler L, Sandbank J, Arenzana-Seisdedos F, Magerus A, Caruz A, Fujii N, Nagler A, Lahav M, Szyper-Kravitz M, Zipori D, Lapidot T. Induction of the chemokine stromal-derived factor-1 following DNA damage improves human stem cell function. J Clin Invest. 2000; 106:1331–1339. DOI: 10.1172/JCI10329. PMID: 11104786. PMCID: 381461.
Article8. Wysoczynski M, Reca R, Ratajczak J, Kucia M, Shirvaikar N, Honczarenko M, Mills M, Wanzeck J, Janowska-Wieczorek A, Ratajczak MZ. Incorporation of CXCR4 into membrane lipid rafts primes homing-related responses of hematopoietic stem/progenitor cells to an SDF-1 gradient. Blood. 2005; 105:40–48. DOI: 10.1182/blood-2004-04-1430.
Article9. Broxmeyer H. Enhancing the efficacy of engraftment of cord blood for hematopoietic cell transplantation. Transfus Apher Sci. 2016; 54:364–372. DOI: 10.1016/j.transci.2016.05.013. PMID: 27211041. PMCID: 4899263.
Article10. Broxmeyer HE, Farag SS, Rocha V. Cord Blood Hematopoietic Cell Transplantation. Forman SJ, Negrin RS, Antin JH, Appelbaum FR, editors. Thomas’ hematopoietic cell transplantation. Oxford: John Wiley & Sons;2016. p. 437–455. DOI: 10.1002/9781118416426.ch39.
Article11. Guo B, Huang X, Cooper S, Broxmeyer HE. Glucocorticoid hormone-induced chromatin remodeling enhances human hematopoietic stem cell homing and engraftment. Nat Med. 2017; 23:424–428. DOI: 10.1038/nm.4298. PMID: 28263313. PMCID: 5408457.
Article12. Huang X, Guo B, Liu S, Wan J, Broxmeyer HE. Neutralizing negative epigenetic regulation by HDAC5 enhances human haematopoietic stem cell homing and engraftment. Nat Commun. 2018; 9:2741. DOI: 10.1038/s41467-018-05178-5. PMID: 30013077. PMCID: 6048146.
Article13. Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008; 455:64–71. DOI: 10.1038/nature07242. PMID: 18668037. PMCID: 2745094.
Article14. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004; 116:281–297. DOI: 10.1016/S0092-8674(04)00045-5. PMID: 14744438.15. Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foà R, Schliwka J, Fuchs U, Novosel A, Müller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, Choksi R, De Vita G, Frezzetti D, Trompeter HI, Hornung V, Teng G, Hartmann G, Palkovits M, Di Lauro R, Wernet P, Macino G, Rogler CE, Nagle JW, Ju J, Papavasiliou FN, Benzing T, Lichter P, Tam W, Brownstein MJ, Bosio A, Borkhardt A, Russo JJ, Sander C, Zavolan M, Tuschl T. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007; 129:1401–1414. DOI: 10.1016/j.cell.2007.04.040. PMID: 17604727. PMCID: 2681231.
Article16. Lee MR, Kim JS, Kim KS. miR-124a is important for migratory cell fate transition during gastrulation of human embryonic stem cells. Stem cells. 2010; 28:1550–1559. DOI: 10.1002/stem.490. PMID: 20665740.
Article17. Houshmand M, Nakhlestani Hagh M, Soleimani M, Hamidieh AA, Abroun S, Nikougoftar Zarif M. MicroRNA microarray profiling during megakaryocyte differentiation of cord blood CD133+ hematopoietic stem cells. Cell J. 2018; 20:195–203. PMID: 29633597. PMCID: 5893291.18. Meng X, Sun B, Xue M, Xu P, Hu F, Xiao Z. Comparative analysis of microRNA expression in human mesenchymal stem cells from umbilical cord and cord blood. Genomics. 2016; 107:124–131. DOI: 10.1016/j.ygeno.2016.02.006. PMID: 26921857.
Article19. Merkerova M, Vasikova A, Belickova M, Bruchova H. MicroRNA expression profiles in umbilical cord blood cell lineages. Stem Cells Dev. 2010; 19:17–26. DOI: 10.1089/scd.2009.0071.
Article20. Peled A, Petit I, Kollet O, Magid M, Ponomaryov T, Byk T, Nagler A, Ben-Hur H, Many A, Shultz L, Lider O, Alon R, Zipori D, Lapidot T. Dependence of human stem cell engraftment and repopulation of NOD/SCID mice on CXCR4. Science. 1999; 283:845–848. DOI: 10.1126/science.283.5403.845. PMID: 9933168.
Article21. Bluteau O, Langlois T, Rivera-Munoz P, Favale F, Rameau P, Meurice G, Dessen P, Solary E, Raslova H, Mercher T, Debili N, Vainchenker W. Developmental changes in human megakaryopoiesis. J Thromb Haemost. 2013; 11:1730–1741. DOI: 10.1111/jth.12326. PMID: 23782903.
Article22. Ferrer-Marin F, Gutti R, Liu ZJ, Sola-Visner M. MiR-9 contributes to the developmental differences in CXCR-4 expression in human megakaryocytes. J Thromb Haemost. 2014; 12:282–285. DOI: 10.1111/jth.12469. PMID: 24738139. PMCID: 3989549.
Article23. Lu J, Luo H, Liu X, Peng Y, Zhang B, Wang L, Xu X, Peng X, Li G, Tian W, He ML, Kung H, Li XP. miR-9 targets CXCR4 and functions as a potential tumor suppressor in nasopharyngeal carcinoma. Carcinogenesis. 2014; 35:554–563. DOI: 10.1093/carcin/bgt354.
Article24. Yu T, Liu K, Wu Y, Fan J, Chen J, Li C, Yang Q, Wang Z. MicroRNA-9 inhibits the proliferation of oral squamous cell carcinoma cells by suppressing expression of CXCR4 via the Wnt/β-catenin signaling pathway. Oncogene. 2014; 33:5017–5027. DOI: 10.1038/onc.2013.448.
Article25. Chatterjee S, Behnam Azad B, Nimmagadda S. The intricate role of CXCR4 in cancer. Adv Cancer Res. 2014; 124:31–82. DOI: 10.1016/B978-0-12-411638-2.00002-1. PMID: 25287686. PMCID: 4322894.
Article26. Selcuklu SD, Donoghue MT, Rehmet K, de Souza Gomes M, Fort A, Kovvuru P, Muniyappa MK, Kerin MJ, Enright AJ, Spillane C. MicroRNA-9 inhibition of cell proliferation and identification of novel miR-9 targets by transcriptome profiling in breast cancer cells. J Biol Chem. 2012; 287:29516–29528. DOI: 10.1074/jbc.M111.335943. PMID: 22761433. PMCID: 3436132.
Article27. Sharma S, Gurudutta G. Epigenetic regulation of hematopoietic stem cells. Int J Stem Cells. 2016; 9:36–43. DOI: 10.15283/ijsc.2016.9.1.36. PMID: 27426084. PMCID: 4961102.
Article28. Guo B, Huang X, Lee MR, Lee SA, Broxmeyer HE. Antagonism of PPAR-γ signaling expands human hematopoietic stem and progenitor cells by enhancing glycolysis. Nat Med. 2018; 24:360–367. DOI: 10.1038/nm.4477. PMID: 29377004. PMCID: 5875416.
Article29. Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005; 33:e179. DOI: 10.1093/nar/gni178. PMID: 16314309. PMCID: 1292995.
Article30. Lee MR, Prasain N, Chae HD, Kim YJ, Mantel C, Yoder MC, Broxmeyer HE. Epigenetic regulation of NANOG by miR-302 cluster-MBD2 completes induced pluripotent stem cell reprogramming. Stem cells. 2013; 31:666–681. DOI: 10.1002/stem.1302.
Article31. Barker JN, Wagner JE. Umbilical cord blood transplantation: current practice and future innovations. Crit Rev Oncol Hematol. 2003; 48:35–43. DOI: 10.1016/S1040-8428(03)00092-1. PMID: 14585482.
Article32. Jaroscak J, Goltry K, Smith A, Waters-Pick B, Martin PL, Driscoll TA, Howrey R, Chao N, Douville J, Burhop S, Fu P, Kurtzberg J. Augmentation of umbilical cord blood (UCB) transplantation with ex vivo-expanded UCB cells: results of a phase 1 trial using the AastromReplicell System. Blood. 2003; 101:5061–5067. DOI: 10.1182/blood-2001-12-0290. PMID: 12595310.
Article33. Marchese A, Raiborg C, Santini F, Keen JH, Stenmark H, Benovic JL. The E3 ubiquitin ligase AIP4 mediates ubiquitination and sorting of the G protein-coupled receptor CXCR4. Dev Cell. 2003; 5:709–722. DOI: 10.1016/S1534-5807(03)00321-6. PMID: 14602072.
Article34. Saini V, Marchese A, Majetschak M. CXC chemokine receptor 4 is a cell surface receptor for extracellular ubiquitin. J Biol Chem. 2010; 285:15566–15576. DOI: 10.1074/jbc.M110.103408. PMID: 20228059. PMCID: 2865327.
Article35. Lapham CK, Romantseva T, Petricoin E, King LR, Manischewitz J, Zaitseva MB, Golding H. CXCR4 heterogeneity in primary cells: possible role of ubiquitination. J Leukoc Biol. 2002; 72:1206–1214. PMID: 12488503.36. Ohno N, Kajiume T, Sera Y, Sato T, Kobayashi M. Short-term culture of umbilical cord blood-derived CD34 cells enhances engraftment into NOD/SCID mice through increased CXCR4 expression. Stem Cells Dev. 2009; 18:1221–1226. DOI: 10.1089/scd.2008.0298.
Article37. Kollet O, Spiegel A, Peled A, Petit I, Byk T, Hershkoviz R, Guetta E, Barkai G, Nagler A, Lapidot T. Rapid and efficient homing of human CD34(+)CD38(-/low)CXCR4(+) stem and progenitor cells to the bone marrow and spleen of NOD/SCID and NOD/SCID/B2m(null) mice. Blood. 2001; 97:3283–3291. DOI: 10.1182/blood.V97.10.3283. PMID: 11342460.
Article38. He L, Zhang L, Wang M, Wang W. miR-9 functions as a tumor inhibitor of cell proliferation in epithelial ovarian cancer through targeting the SDF-1/CXCR4 pathway. Exp Ther Med. 2017; 13:1203–1208. DOI: 10.3892/etm.2017.4118. PMID: 28413458. PMCID: 5377313.
Article39. Brunstein CG, Barker JN, Weisdorf DJ, DeFor TE, Miller JS, Blazar BR, McGlave PB, Wagner JE. Umbilical cord blood transplantation after nonmyeloablative conditioning: impact on transplantation outcomes in 110 adults with hematologic disease. Blood. 2007; 110:3064–3070. DOI: 10.1182/blood-2007-04-067215. PMID: 17569820. PMCID: 2018678.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Atorvastatin Pretreatment Ameliorates Mesenchymal Stem Cell Migration through miR-146a/CXCR4 Signaling
- Isoorientin Suppresses Invasion of Breast and Colon Cancer Cells by Inhibition of CXC Chemokine Receptor 4 Expression
- Human Umbilical Cord Blood CD34-Positive Cells as Predictors of the Incidence and Short-Term Outcome of Neonatal Hypoxic-Ischemic Encephalopathy: A Pilot Study
- MicroRNA-765 is upregulated in myelodysplastic syndromes and induces apoptosis via PLP2 inhibition in leukemia cells
- Mitochondria Activity and CXCR4 Collaboratively Promote the Differentiation of CD11c + B Cells Induced by TLR9 in Lupus