J Vet Sci.
2018 Nov;19(6):798-807. 10.4142/jvs.2018.19.6.798.
Benefits of procyanidins on gut microbiota in Bama minipigs and implications in replacing antibiotics
- Affiliations
-
- 1Key Laboratory of Veterinary Biotechnology, School of Agriculture and Biology, Shanghai Jiao Tong University, Shanghai 200240, China. lcui@sjtu.edu.cn
- KMID: 2427028
- DOI: http://doi.org/10.4142/jvs.2018.19.6.798
Abstract
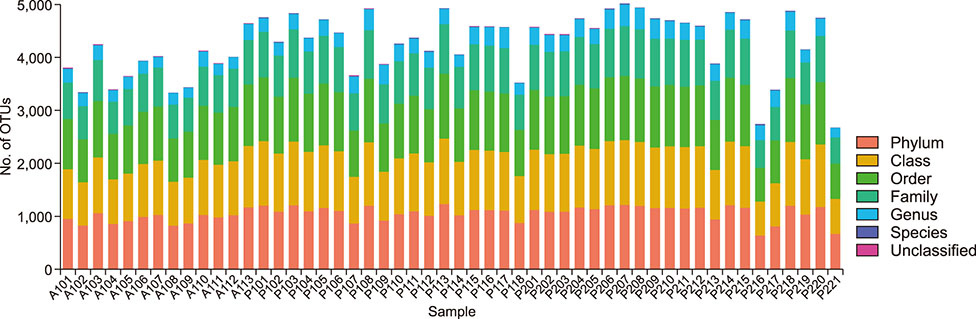
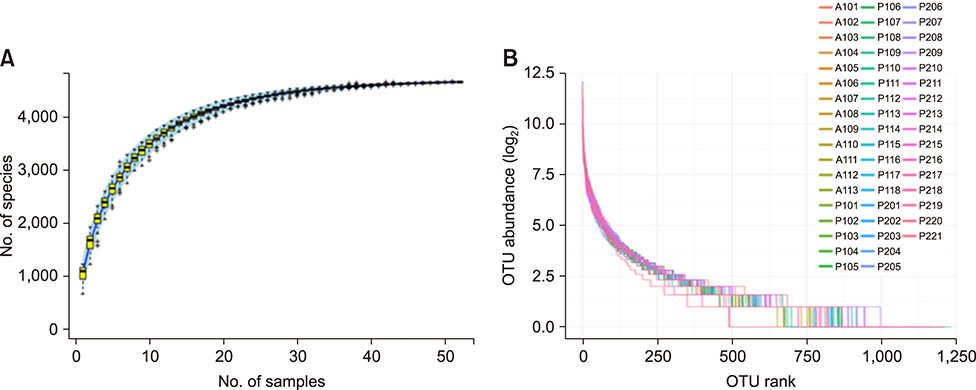
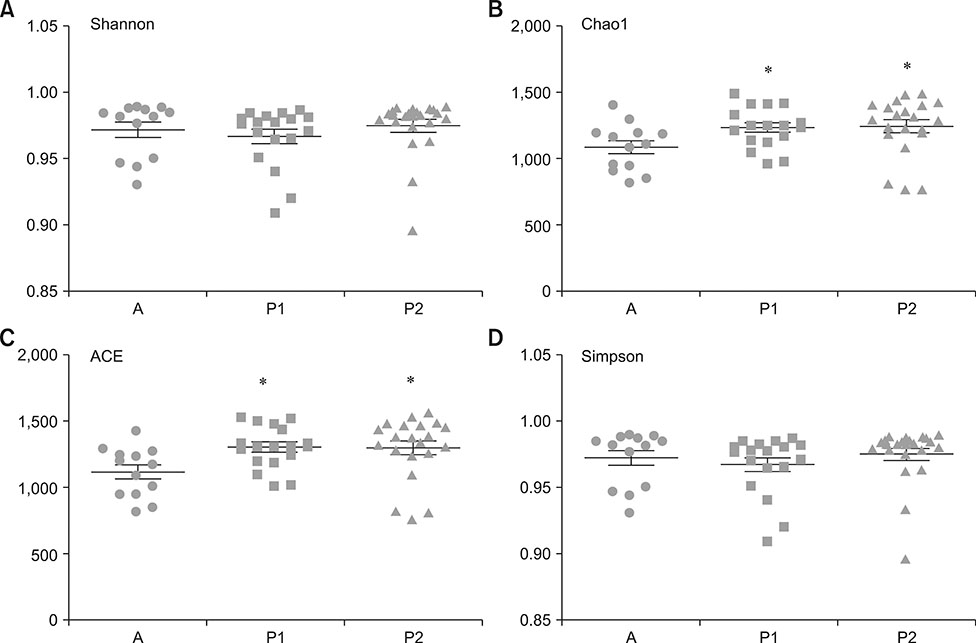
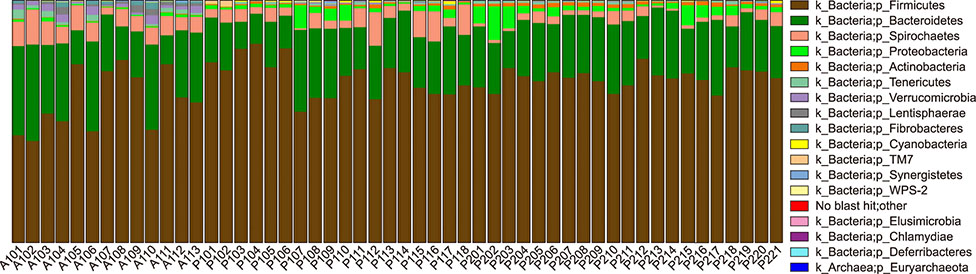
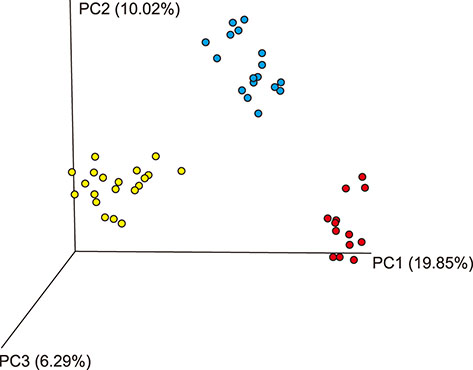
- Several studies have reported the effect of absorption of procyanidins and their contribution to the small intestine. However, differences between dietary interventions of procyanidins and interventions via antibiotic feeding in pigs are rarely reported. Following 16S rRNA gene Illumina MiSeq sequencing, we observed that both procyanidin administration for 2 months (procyanidin-1 group) and continuous antibiotic feeding for 1 month followed by procyanidin for 1 month (procyanidin-2 group) increased the number of operational taxonomic units, as well as the Chao 1 and ACE indices, compared to those in pigs undergoing antibiotic administration for 2 months (antibiotic group). The genera Fibrobacter and Spirochaete were more abundant in the antibiotic group than in the procyanidin-1 and procyanidin-2 groups. Principal component analysis revealed clear separations among the three groups. Additionally, using the online Molecular Ecological Network Analyses pipeline, three co-occurrence networks were constructed; Lactobacillus was in a co-occurrence relationship with Trichococcus and Desulfovibrio and a co-exclusion relationship with Bacillus and Spharerochaeta. Furthermore, metabolic function analysis by phylogenetic investigation of communities by reconstruction of unobserved states demonstrated modulation of pathways involved in the metabolism of carbohydrates, amino acids, energy, and nucleotides. These data suggest that procyanidin influences the gut microbiota and the intestinal metabolic function to produce beneficial effects on metabolic homeostasis.
MeSH Terms
-
Absorption
Amino Acids
Anti-Bacterial Agents*
Bacillus
Carbohydrates
Desulfovibrio
Fibrobacter
Gastrointestinal Microbiome*
Genes, rRNA
Homeostasis
Intestine, Small
Lactobacillus
Metabolism
Nucleotides
Principal Component Analysis
Proanthocyanidins*
Swine
Swine, Miniature*
Amino Acids
Anti-Bacterial Agents
Carbohydrates
Nucleotides
Proanthocyanidins
Figure
Reference
-
1. Appeldoorn MM, Vincken JP, Gruppen H, Hollman PC. Procyanidin dimers A1, A2, and B2 are absorbed without conjugation or methylation from the small intestine of rats. J Nutr. 2009; 139:1469–1473.
Article2. Broadhurst MJ, Ardeshir A, Kanwar B, Mirpuri J, Gundra UM, Leung JM, Wiens KE, Vujkovic-Cvijin I, Kim CC, Yarovinsky F, Lerche NW, McCune JM, Loke P. Therapeutic helminth infection of macaques with idiopathic chronic diarrhea alters the inflammatory signature and mucosal microbiota of the colon. PLoS Pathog. 2012; 8:e1003000.
Article3. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010; 7:335–336.
Article4. Carmody RN, Gerber GK, Luevano JM Jr, Gatti DM, Somes L, Svenson KL, Turnbaugh PJ. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe. 2015; 17:72–84.
Article5. Cho I, Yamanishi S, Cox L, Methé BA, Zavadil J, Li K, Gao Z, Mahana D, Raju K, Teitler I, Li H, Alekseyenko AV, Blaser MJ. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. 2012; 488:621–626.
Article6. Claesson MJ, Jeffery IB, Conde S, Power SE, O'Connor EM, Cusack S, Harris HM, Coakley M, Lakshminarayanan B, O'Sullivan O, Fitzgerald GF, Deane J, O'Connor M, Harnedy N, O'Connor K, O'Mahony D, van Sinderen D, Wallace M, Brennan L, Stanton C, Marchesi JR, Fitzgerald AP, Shanahan F, Hill C, Ross RP, O'Toole PW. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012; 488:178–184.
Article7. Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012; 148:1258–1270.
Article8. Cox A, Dixon P, Smith S. Histopathological lesions associated with equine periodontal disease. Vet J. 2012; 194:386–391.
Article9. De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010; 107:14691–14696.
Article10. Emoto T, Yamashita T, Sasaki N, Hirota Y, Hayashi T, So A, Kasahara K, Yodoi K, Matsumoto T, Mizoguchi T, Ogawa W, Hirata K. Analysis of gut microbiota in coronary artery disease patients: a possible link between gut microbiota and coronary artery disease. J Atheroscler Thromb. 2016; 23:908–921.
Article11. Faria A, Calhau C, de Freitas V, Mateus N. Procyanidins as antioxidants and tumor cell growth modulators. J Agric Food Chem. 2006; 54:2392–2397.
Article12. Faust K, Sathirapongsasuti JF, Izard J, Segata N, Gevers D, Raes J, Huttenhower C. Microbial co-occurrence relationships in the human microbiome. PLoS Comput Biol. 2012; 8:e1002606.
Article13. Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes. 2012; 3:289–306.
Article14. Frese SA, Parker K, Calvert CC, Mills DA. Diet shapes the gut microbiome of pigs during nursing and weaning. Microbiome. 2015; 3:28.
Article15. Gonçalves C, Dinis T, Batista MT. Antioxidant properties of proanthocyanidins of Uncaria tomentosa bark decoction: a mechanism for anti-inflammatory activity. Phytochemistry. 2005; 66:89–98.
Article16. Gulgun M, Erdem O, Oztas E, Kesik V, Balamtekin N, Vurucu S, Kul M, Kismet E, Koseoglu V. Proanthocyanidin prevents methotrexate-induced intestinal damage and oxidative stress. Exp Toxicol Pathol. 2010; 62:109–115.
Article17. Jerez M, Deive FJ, Sineiro J, Núñez MJ. Antioxidant activity of pine bark procyanidins in bulk corn oil and corn oil-in-water emulsions. Eur J Lipid Sci Tech. 2011; 113:1402–1411.
Article18. Kashyap PC, Marcobal A, Ursell LK, Larauche M, Duboc H, Earle KA, Sonnenburg ED, Ferreyra JA, Higginbottom SK, Million M, Tache Y, Pasricha PJ, Knight R, Farrugia G, Sonnenburg JL. Complex interactions among diet, gastrointestinal transit, and gut microbiota in humanized mice. Gastroenterology. 2013; 144:967–977.
Article19. Kim DS, Kim MS, Kang SW, Sung HY, Kang YH. Pine bark extract enzogenol attenuated tumor necrosis factor-α-induced endothelial cell adhesion and monocyte transmigration. J Agric Food Chem. 2010; 58:7088–7095.
Article20. Kim YJ, Park HJ, Yoon SH, Kim MJ, Leem KH, Chung JH, Kim HK. Anticancer effects of oligomeric proanthocyanidins on human colorectal cancer cell line, SNU-C4. World J Gastroenterol. 2005; 11:4674–4678.
Article21. Kimura Y, Sumiyoshi M. French maritime pine bark (Pinus maritima Lam.) extract (Flavangenol®) prevents chronic UVB radiation-induced skin damage and carcinogenesis in melanin-possessing hairless mice. Photochem Photobiol. 2010; 86:955–963.
Article22. Kusuda M, Inada K, Ogawa TO, Yoshida T, Shiota S, Tsuchiya T, Hatano T. Polyphenolic constituent structures of Zanthoxylum piperitum fruit and the antibacterial effects of its polymeric procyanidin on methicillin-resistant Staphylococcus aureus. Biosci Biotechnol Biochem. 2006; 70:1423–1431.
Article23. Li J, Liu Y, Zhang JW, Wei H, Yang L. Characterization of hepatic drug-metabolizing activities of Bama miniature pigs (Sus scrofa domestica): comparison with human enzyme analogs. Comp Med. 2006; 56:286–290.24. Li K, Li Q, Li J, Zhang T, Han Z, Gao D, Zheng F. Antitumor activity of the procyanidins from Pinus koraiensis Bark on mice bearing U14 cervical cancer. Yakugaku Zasshi. 2007; 127:1145–1151.
Article25. Lin A, Bik EM, Costello EK, Dethlefsen L, Haque R, Relman DA, Singh U. Distinct distal gut microbiome diversity and composition in healthy children from Bangladesh and the United States. PLoS One. 2013; 8:e53838.
Article26. Liu Y, Zeng BH, Shang HT, Cen YY, Wei H. Bama miniature pigs (Sus scrofa domestica) as a model for drug evaluation for humans: comparison of in vitro metabolism and in vivo pharmacokinetics of lovastatin. Comp Med. 2008; 58:580–587.27. Mantena SK, Baliga MS, Katiyar SK. Grape seed proanthocyanidins induce apoptosis and inhibit metastasis of highly metastatic breast carcinoma cells. Carcinogenesis. 2006; 27:1682–1691.
Article28. Masumoto S, Terao A, Yamamoto Y, Mukai T, Miura T, Shoji T. Non-absorbable apple procyanidins prevent obesity associated with gut microbial and metabolomic changes. Sci Rep. 2016; 6:31208.
Article29. McArdle BH, Anderson MJ. Fitting multivariate models to community data: a comment on distance-based redundancy analysis. Ecology. 2001; 82:290–297.
Article30. Ming DS, López A, Hillhouse BJ, French CJ, Hudson JB, Towers GH. Bioactive constituents from Iryanthera megistophylla. J Nat Prod. 2002; 65:1412–1416.31. Newman ME. Modularity and community structure in networks. Proc Natl Acad Sci U S A. 2006; 103:8577–8582.
Article32. Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, Peng Y, Zhang D, Jie Z, Wu W, Qin Y, Xue W, Li J, Han L, Lu D, Wu P, Dai Y, Sun X, Li Z, Tang A, Zhong S, Li X, Chen W, Xu R, Wang M, Feng Q, Gong M, Yu J, Zhang Y, Zhang M, Hansen T, Sanchez G, Raes J, Falony G, Okuda S, Almeida M, LeChatelier E, Renault P, Pons N, Batto JM, Zhang Z, Chen H, Yang R, Zheng W, Li S, Yang H, Wang J, Ehrlich SD, Nielsen R, Pedersen O, Kristiansen K, Wang J. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012; 490:55–60.
Article33. Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013; 41:D590–D596.
Article34. Relman DA. Microbiology: learning about who we are. Nature. 2012; 486:194–195.35. Rios LY, Bennett RN, Lazarus SA, Rémésy C, Scalbert A, Williamson G. Cocoa procyanidins are stable during gastric transit in humans. Am J Clin Nutr. 2002; 76:1106–1110.
Article36. Rohdewald P. A review of the French maritime pine bark extract (Pycnogenol), a herbal medication with a diverse clinical pharmacology. Int J Clin Pharmacol Ther. 2002; 40:158–168.
Article37. Scheperjans F, Aho V, Pereira PA, Koskinen K, Paulin L, Pekkonen E, Haapaniemi E, Kaakkola S, Eerola-Rautio J, Pohja M, Kinnunen E, Murros K, Auvinen P. Gut microbiota are related to Parkinson's disease and clinical phenotype. Mov Disord. 2015; 30:350–358.
Article38. Scott KP, Gratz SW, Sheridan PO, Flint HJ, Duncan SH. The influence of diet on the gut microbiota. Pharmacol Res. 2013; 69:52–60.
Article39. Shang HT, Niu R, Wei H, Huang ZB, Gan S, Wang A, Zeng Y. Genetic analysis of 35 microsatellite loci in three miniature pig breeds. Hereditas. 2001; 23:17–20.40. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003; 13:2498–2504.
Article41. Spencer JP, Schroeter H, Rechner AR, Rice-Evans C. Bioavailability of flavan-3-ols and procyanidins: gastrointestinal tract influences and their relevance to bioactive forms in vivo. Antioxid Redox Signal. 2001; 3:1023–1039.
Article42. Tilocca B, Burbach K, Heyer CME, Hoelzle LE, Mosenthin R, Stefanski V, Camarinha-Silva A, Seifert J. Dietary changes in nutritional studies shape the structural and functional composition of the pigs' fecal microbiome–from days to weeks. Microbiome. 2017; 5:144.
Article43. Tremaroli V, Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012; 489:242–249.
Article44. Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. A core gut microbiome in obese and lean twins. Nature. 2009; 457:480–484.
Article45. Tzounis X, Rodriguez-Mateos A, Vulevic J, Gibson GR, Kwik-Uribe C, Spencer JP. Prebiotic evaluation of cocoa-derived flavanols in healthy humans by using a randomized, controlled, double-blind, crossover intervention study. Am J Clin Nutr. 2011; 93:62–72.
Article46. Visser MB, Ellen RP. New insights into the emerging role of oral spirochaetes in periodontal disease. Clin Microbiol Infect. 2011; 17:502–512.
Article47. Warton DI, Wright ST, Wang Y. Distance-based multivariate analyses confound location and dispersion effects. Method Ecol Evol. 2012; 3:89–101.
Article48. Wu S, Li RW, Li W, Beshah E, Dawson HD, Urban JF Jr. Worm burden-dependent disruption of the porcine colon microbiota by Trichuris suis infection. PLoS One. 2012; 7:e35470.49. Xiao Y, Li K, Xiang Y, Zhou W, Gui G, Yang H. The fecal microbiota composition of boar Duroc, Yorkshire, Landrace and Hampshire pigs. Asian-Australas J Anim Sci. 2017; 30:1456–1463.
Article50. Zaura E, Keijser BJ, Huse SM, Crielaard W. Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol. 2009; 9:259.
Article