Cancer Res Treat.
2018 Oct;50(4):1203-1213. 10.4143/crt.2017.538.
CCR6 Is a Predicting Biomarker of Radiosensitivity and Potential Target of Radiosensitization in Rectal Cancer
- Affiliations
-
- 1State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Guangzhou, China. gaoyh1962@126.com, xiaoww@sysucc.org.cn
- 2Department of Radiation Oncology, Sun Yat-sen University Cancer Center, Guangzhou, China.
- 3Department of Colorectal Surgery, Sun Yat-sen University Cancer Center, Guangzhou, China.
- KMID: 2424793
- DOI: http://doi.org/10.4143/crt.2017.538
Abstract
- PURPOSE
This study aimed to explore the functions and mechanisms of C-C motif chemokine receptor 6 (CCR6), a gene associated with progression and metastasis of colorectal cancer (CRC), in radiosensitivity of rectal cancer (RC).
MATERIALS AND METHODS
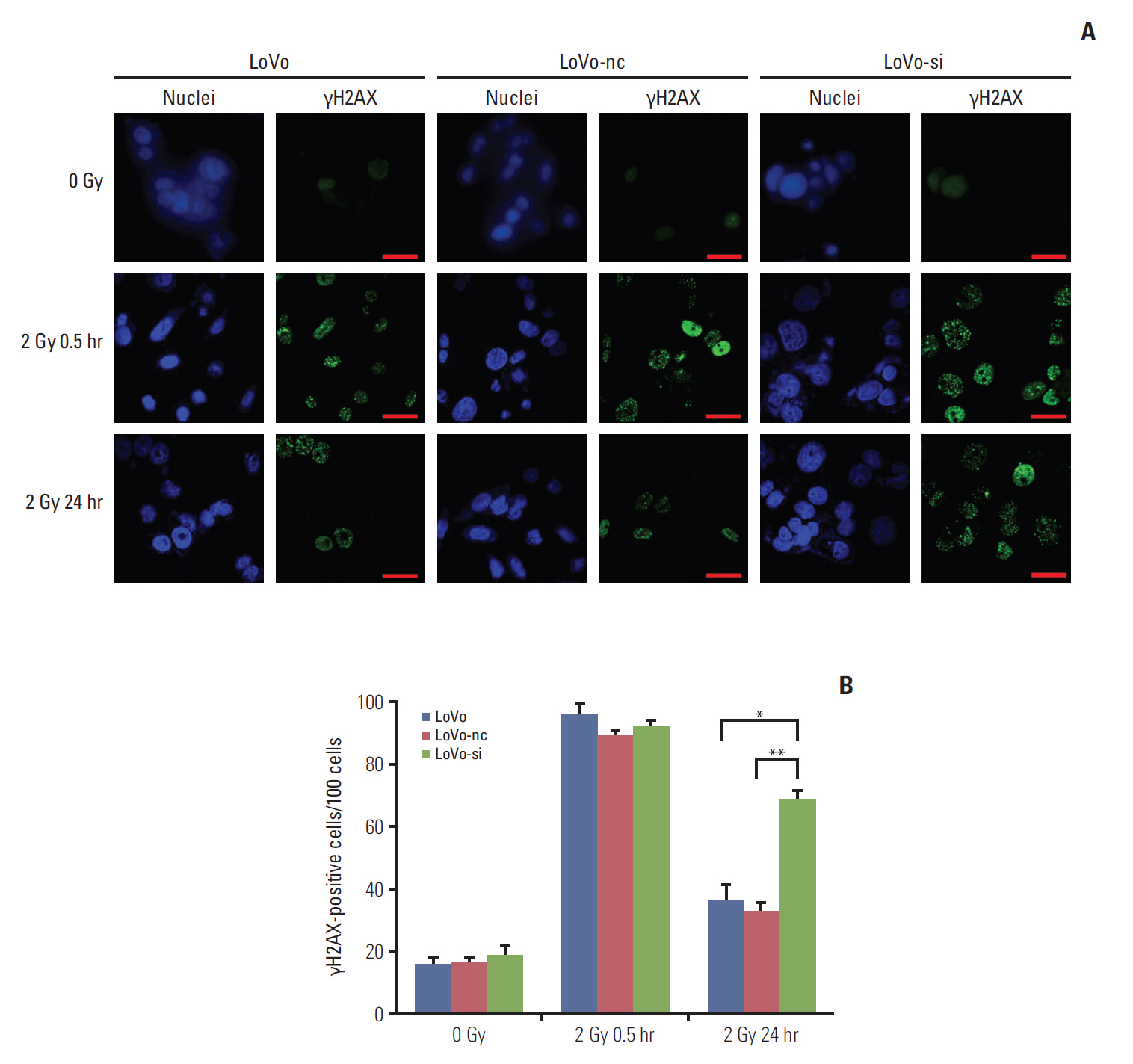
RNA sequencing and immunohistochemical analysis on CCR6 expression were performed in pretreatment tissues of RC patients exhibiting different therapeutic effects of radiotherapy. Colonogenic survival assay was conducted in different CRC cell lines to assess their radiosensitivity. And the impact of CCR6 expression on radiosensitivity was validated through RNA interference. The DNA damage repair (DDR) abilities of cell lines with different CCR6 expression were evaluated through immunofluorescence-based γH2AX quantification.
RESULTS
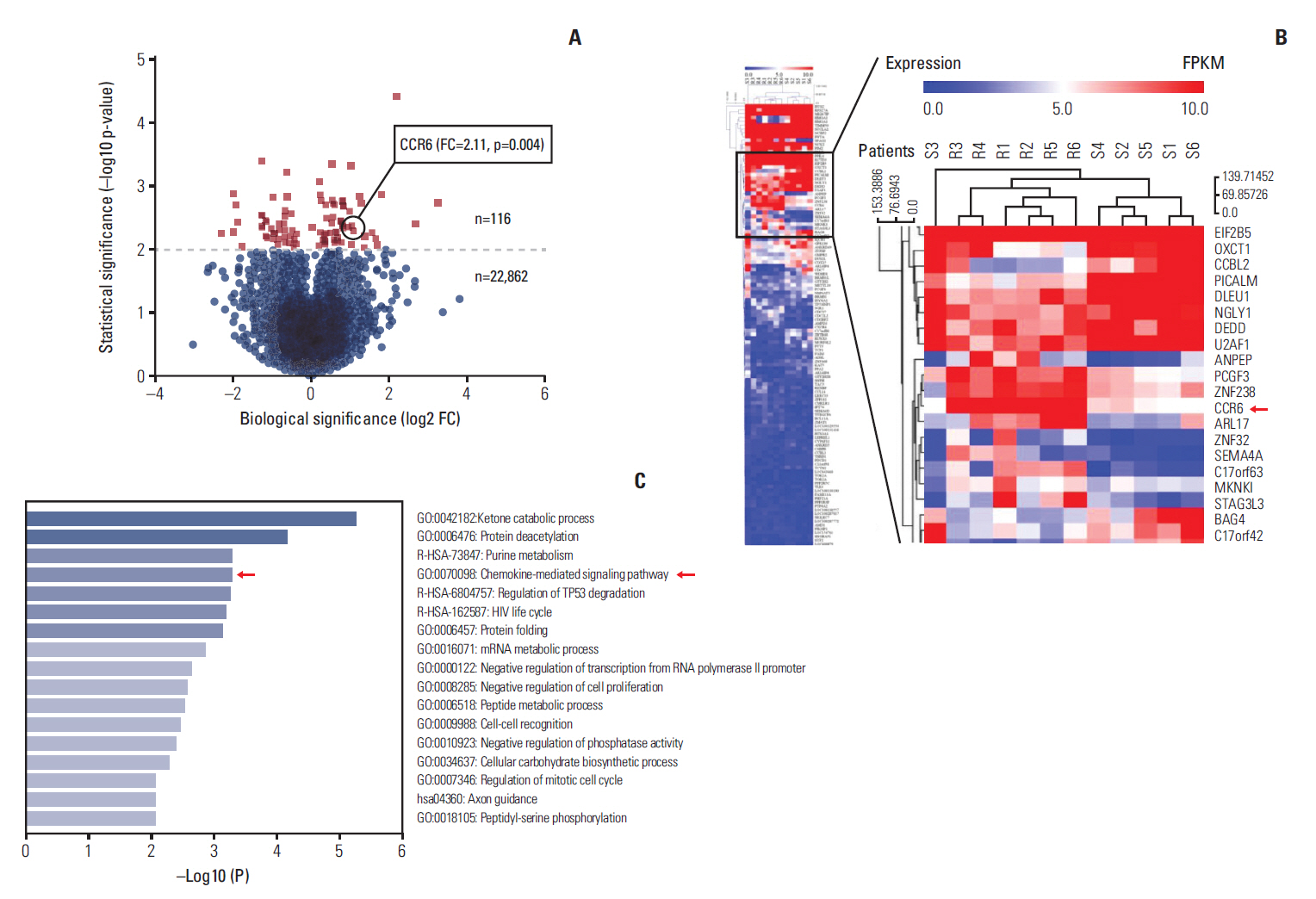
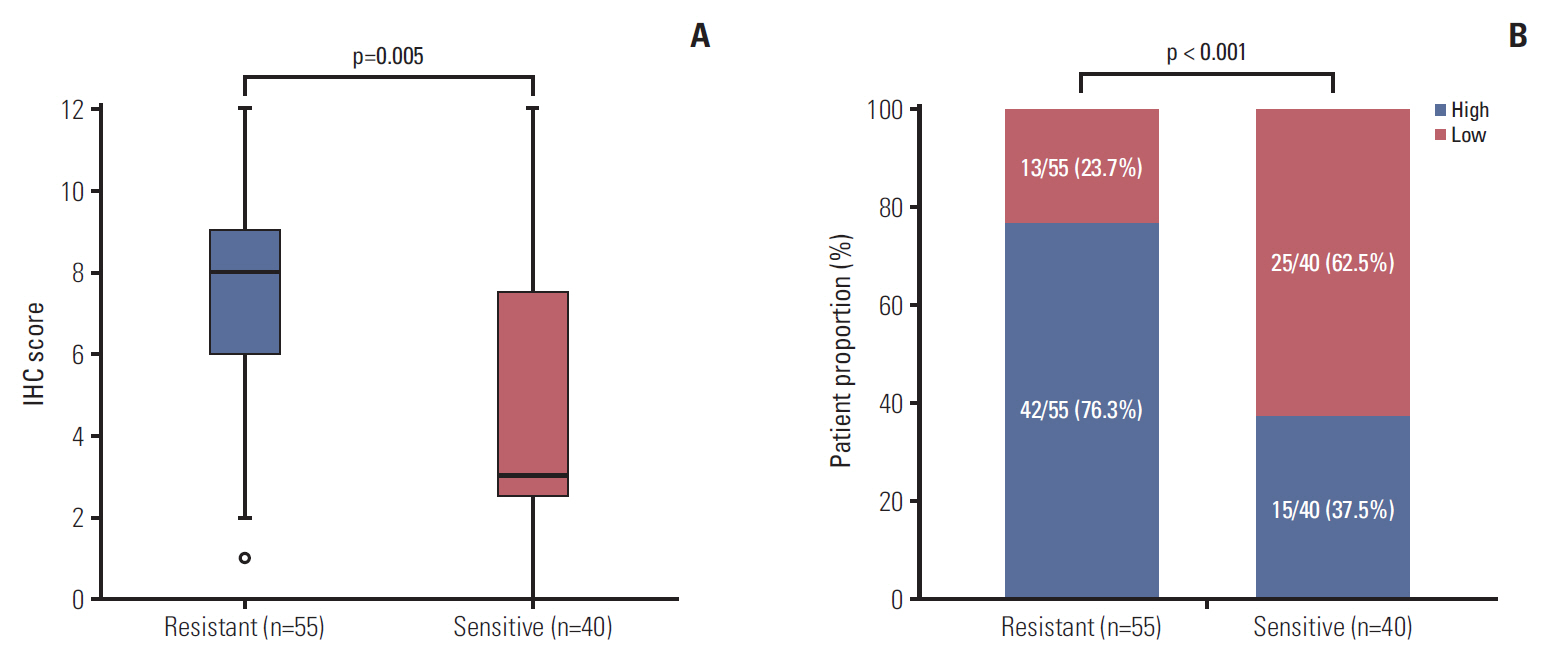
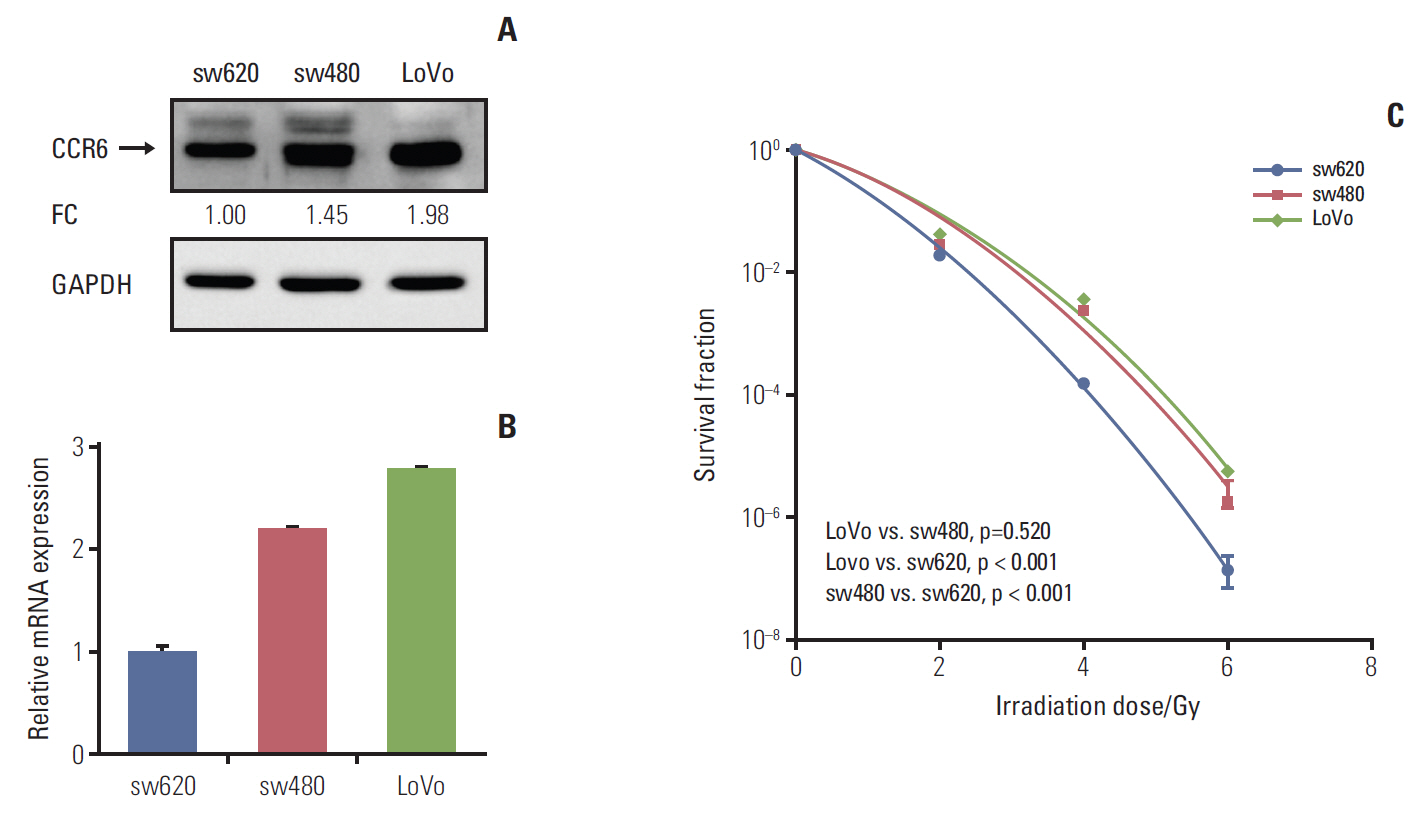
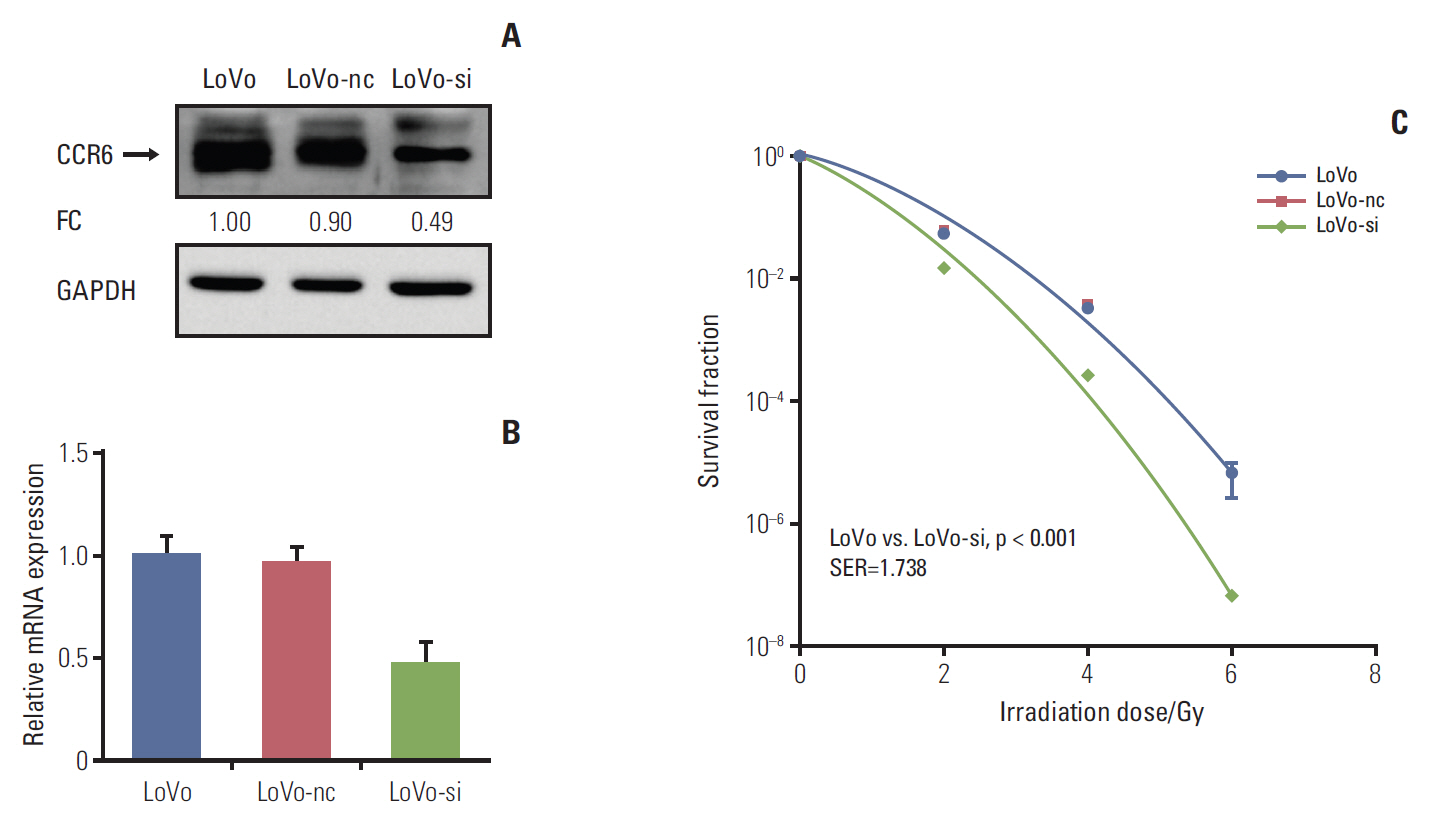
The CCR6 mRNA level was higher in patients without pathologic complete remission (pCR) than in those with pCR (fold changed, 2.11; p=0.004). High-level expression of CCR6 protein was more common in the bad responders than in the good responders (76.3% vs. 37.5%, p < 0.001). The CRC cell lines with higher CCR6 expression (LoVo and sw480) appeared to be more radioresistant, compared with the sw620 cell line which had lower CCR6 expression. CCR6 knockdown made the LoVo cells more sensitive to ionizing radiation (sensitization enhancement ratio, 1.738; p < 0.001), and decreased their DDR efficiency.
CONCLUSION
CCR6 might affect the RC radiosensitivity through DDR process. These findings supported CCR6 as a predicting biomarker of radiosensitivity and a potential target of radiosensitization for RC patients.
Keyword
MeSH Terms
Figure
Reference
-
References
1. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016; 66:115–32.
Article2. Park HJ, Cho S, Kim Y. Patterns of rectal cancer radiotherapy adopting evidence-based medicine: an analysis of the national database from 2005 to 2016. Cancer Res Treat. 2018; 50:975–83.
Article3. Fokas E, Liersch T, Fietkau R, Hohenberger W, Beissbarth T, Hess C, et al. Tumor regression grading after preoperative chemoradiotherapy for locally advanced rectal carcinoma revisited: updated results of the CAO/ARO/AIO-94 trial. J Clin Oncol. 2014; 32:1554–62.
Article4. Trakarnsanga A, Gonen M, Shia J, Nash GM, Temple LK, Guillem JG, et al. Comparison of tumor regression grade systems for locally advanced rectal cancer after multimodality treatment. J Natl Cancer Inst. 2014; 106:dju248.
Article5. Morgan MA, Lawrence TS. Molecular pathways: overcoming radiation resistance by targeting DNA damage response pathways. Clin Cancer Res. 2015; 21:2898–904.
Article6. Barker HE, Paget JT, Khan AA, Harrington KJ. The tumour microenvironment after radiotherapy: mechanisms of resistance and recurrence. Nat Rev Cancer. 2015; 15:409–25.
Article7. Zlotnik A, Burkhardt AM, Homey B. Homeostatic chemokine receptors and organ-specific metastasis. Nat Rev Immunol. 2011; 11:597–606.
Article8. Ghadjar P, Coupland SE, Na IK, Noutsias M, Letsch A, Stroux A, et al. Chemokine receptor CCR6 expression level and liver metastases in colorectal cancer. J Clin Oncol. 2006; 24:1910–6.
Article9. Kapur N, Mir H, Clark CE 3rd, Krishnamurti U, Beech DJ, Lillard JW, et al. CCR6 expression in colon cancer is associated with advanced disease and supports epithelial-to-mesenchymal transition. Br J Cancer. 2016; 114:1343–51.
Article10. Nandi B, Pai C, Huang Q, Prabhala RH, Munshi NC, Gold JS. CCR6, the sole receptor for the chemokine CCL20, promotes spontaneous intestinal tumorigenesis. PLoS One. 2014; 9:e97566.
Article11. Frick VO, Rubie C, Keilholz U, Ghadjar P. Chemokine/chemokine receptor pair CCL20/CCR6 in human colorectal malignancy: an overview. World J Gastroenterol. 2016; 22:833–41.
Article12. Hu L, Li X, Liu Q, Xu J, Ge H, Wang Z, et al. UBE2S, a novel substrate of Akt1, associates with Ku70 and regulates DNA repair and glioblastoma multiforme resistance to chemotherapy. Oncogene. 2017; 36:1145–56.
Article13. Ko JC, Chen JC, Wang TJ, Zheng HY, Chen WC, Chang PY, et al. Astaxanthin down-regulates Rad51 expression via inactivation of AKT kinase to enhance mitomycin C-induced cytotoxicity in human non-small cell lung cancer cells. Biochem Pharmacol. 2016; 105:91–100.
Article14. Marampon F, Gravina GL, Zani BM, Popov VM, Fratticci A, Cerasani M, et al. Hypoxia sustains glioblastoma radioresistance through ERKs/DNA-PKcs/HIF-1alpha functional interplay. Int J Oncol. 2014; 44:2121–31.15. Mandric I, Temate-Tiagueu Y, Shcheglova T, Al Seesi S, Zelikovsky A, Mandoiu II. Fast bootstrapping-based estimation of confidence intervals of expression levels and differential expression from RNA-Seq data. Bioinformatics. 2017; 33:3302–4.
Article16. Tripathi S, Pohl MO, Zhou Y, Rodriguez-Frandsen A, Wang G, Stein DA, et al. Meta- and orthogonal integration of influenza "OMICs" data defines a role for UBR4 in virus budding. Cell Host Microbe. 2015; 18:723–35.
Article17. Chen DL, Chen LZ, Lu YX, Zhang DS, Zeng ZL, Pan ZZ, et al. Long noncoding RNA XIST expedites metastasis and modulates epithelial-mesenchymal transition in colorectal cancer. Cell Death Dis. 2017; 8:e3011.
Article18. Cerda MB, Lloyd R, Batalla M, Giannoni F, Casal M, Policastro L. Silencing peroxiredoxin-2 sensitizes human colorectal cancer cells to ionizing radiation and oxaliplatin. Cancer Lett. 2017; 388:312–9.
Article19. Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, et al. Primer3: new capabilities and interfaces. Nucleic Acids Res. 2012; 40:e115.20. Tu Z, Xu B, Qu C, Tao Y, Chen C, Hua W, et al. BRCC3 acts as a prognostic marker in nasopharyngeal carcinoma patients treated with radiotherapy and mediates radiation resistance in vitro. Radiat Oncol. 2015; 10:123.
Article21. Kleiner RE, Verma P, Molloy KR, Chait BT, Kapoor TM. Chemical proteomics reveals a gammaH2AX-53BP1 interaction in the DNA damage response. Nat Chem Biol. 2015; 11:807–14.22. Chin CC, Chen CN, Kuo HC, Shi CS, Hsieh MC, Kuo YH, et al. Interleukin-17 induces CC chemokine receptor 6 expression and cell migration in colorectal cancer cells. J Cell Physiol. 2015; 230:1430–7.
Article23. Maier P, Hartmann L, Wenz F, Herskind C. Cellular pathways in response to ionizing radiation and their targetability for tumor radiosensitization. Int J Mol Sci. 2016; 17:E102.
Article24. Lee JS. Activation of ATM-dependent DNA damage signal pathway by a histone deacetylase inhibitor, trichostatin A. Cancer Res Treat. 2007; 39:125–30.
Article25. Moon SH, Lin L, Zhang X, Nguyen TA, Darlington Y, Waldman AS, et al. Wild-type p53-induced phosphatase 1 dephosphorylates histone variant gamma-H2AX and suppresses DNA double strand break repair. J Biol Chem. 2010; 285:12935–47.26. Zhu Y, Dai B, Zhang H, Shi G, Shen Y, Ye D. Long non-coding RNA LOC572558 inhibits bladder cancer cell proliferation and tumor growth by regulating the AKT-MDM2-p53 signaling axis. Cancer Lett. 2016; 380:369–74.
Article27. Mendoza MC, Er EE, Blenis J. The Ras-ERK and PI3K-mTOR pathways: cross-talk and compensation. Trends Biochem Sci. 2011; 36:320–8.
Article28. Sato A, Sunayama J, Matsuda K, Seino S, Suzuki K, Watanabe E, et al. MEK-ERK signaling dictates DNA-repair gene MGMT expression and temozolomide resistance of stem-like glioblastoma cells via the MDM2-p53 axis. Stem Cells. 2011; 29:1942–51.
Article29. Sun Y. Tumor microenvironment and cancer therapy resistance. Cancer Lett. 2016; 380:205–15.
Article30. Trautmann F, Cojoc M, Kurth I, Melin N, Bouchez LC, Dubrovska A, et al. CXCR4 as biomarker for radioresistant cancer stem cells. Int J Radiat Biol. 2014; 90:687–99.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- FXYD-3 expression in relation to local recurrence of rectal cancer
- Enhanced Radiosensitivity of Tumor Cells Treated with Vanadate in Vitro
- Isotype-Specific Inhibition of Histone Deacetylases: Identification of Optimal Targets for Radiosensitization
- Metastasis-associated protein 1: a druggable target in cancer treatment
- A Case of Rectal Cancer in 12 year Old Boy