J Breast Cancer.
2018 Sep;21(3):233-243. 10.4048/jbc.2018.21.e41.
The Epigenetics of Triple-Negative and Basal-Like Breast Cancer: Current Knowledge
- Affiliations
-
- 1Research Center for Functional Genomics, Biomedicine and Translational Medicine, “Iuliu Hatieganu†University of Medicine and Pharmacy, Cluj-Napoca, Romania.
- 2Faculty of Medicine, “Iuliu Hatieganu†University of Medicine and Pharmacy, Cluj-Napoca, Romania.
- 3Division of Dental Propaedeutics, Aesthetic, Department of Prosthetic Dentistry and Dental Materials, “Iuliu Hatieganu†University of Medicine and Pharmacy, Cluj-Napoca, Romania. irimie.alexandra@umfcluj.ro
- 4MedFUTURE Research Center for Advanced Medicine, “Iuliu Hatieganu†University of Medicine and Pharmacy, Cluj-Napoca, Romania.
- 5Department of Functional Genomics and Experimental Pathology, The Oncology Institute “Prof. Dr. I Chiricutaâ€, Cluj-Napoca, Romania.
- KMID: 2421363
- DOI: http://doi.org/10.4048/jbc.2018.21.e41
Abstract
- Breast cancer has the highest incidence among all malignancies diagnosed in women. Therapies have significantly improved over the years due to extensive molecular and clinical research; in a large number of cases, targeted therapies have provided better prognosis. However, one specific subtype remains elusive to targeted therapies-the triple-negative breast cancer. This immunohistochemically defined subtype is resistant to both endocrine and targeted therapies, leading to its poor prognosis. A field that is of great promise in current cancer research is epigenetics. By studying the epigenetic mechanisms underlying tumorigenesis-DNA methylation, histone modifications, and noncoding RNAs-advances in cancer treatment, diagnosis, and prevention are possible. This review aims to synthesize the epigenetic discoveries that have been made related to the triple-negative breast cancer.
Keyword
MeSH Terms
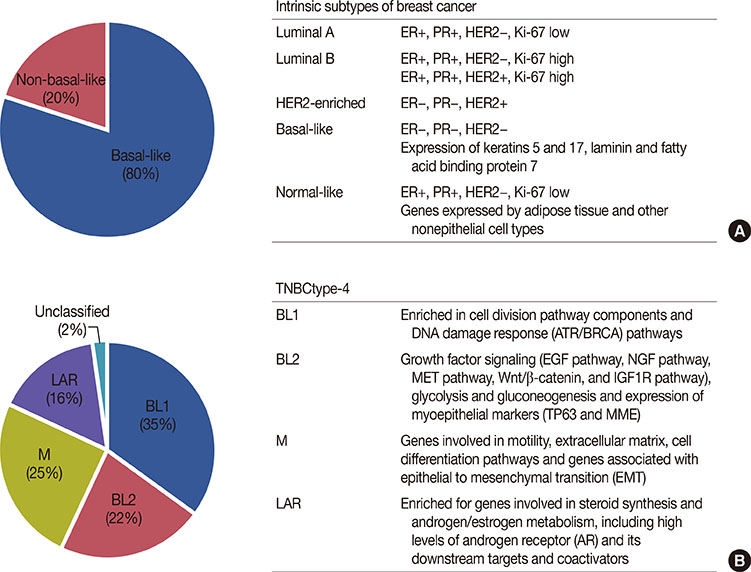
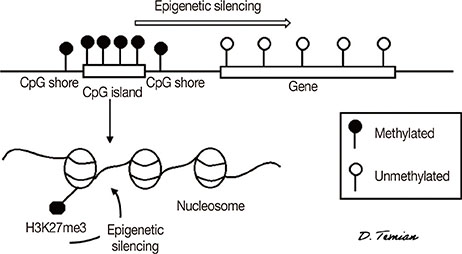
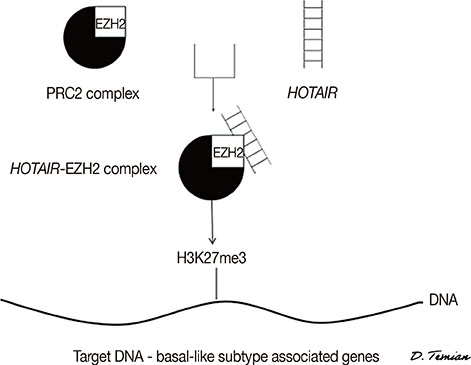
Figure
Reference
-
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018; 68:7–30.
Article2. Gierach GL, Burke A, Anderson WF. Epidemiology of triple negative breast cancers. Breast Dis. 2010; 32:5–24.
Article3. Chiorean R, Braicu C, Berindan-Neagoe I. Another review on triple negative breast cancer: are we on the right way towards the exit from the labyrinth? Breast. 2013; 22:1026–1033.
Article4. Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000; 406:747–752.
Article5. Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001; 98:10869–10874.
Article6. Senkus E, Kyriakides S, Penault-Llorca F, Poortmans P, Thompson A, Zackrisson S, et al. Primary breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2013; 24:Suppl 6. vi7–vi23.
Article7. Lachapelle J, Foulkes WD. Triple-negative and basal-like breast cancer: implications for oncologists. Curr Oncol. 2011; 18:161–164.
Article8. Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010; 363:1938–1948.
Article9. Lehmann BD, Pietenpol JA. Identification and use of biomarkers in treatment strategies for t riple-negative breast cancer subtypes. J Pathol. 2014; 232:142–150.
Article10. Perou CM. Molecular stratification of triple-negative breast cancers. Oncologist. 2011; 16:Suppl 1. 61–70.
Article11. Weigelt B, Baehner FL, Reis-Filho JS. The contribution of gene expression profiling to breast cancer classification, prognostication and prediction: a retrospective of the last decade. J Pathol. 2010; 220:263–280.
Article12. Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011; 121:2750–2767.
Article13. Lehmann BD, Jovanović B, Chen X, Estrada MV, Johnson KN, Shyr Y, et al. Refinement of triple-negative breast cancer molecular subtypes: implications for neoadjuvant chemotherapy selection. PLoS One. 2016; 11:e0157368.
Article14. Burstein MD, Tsimelzon A, Poage GM, Covington KR, Contreras A, Fuqua SA, et al. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin Cancer Res. 2015; 21:1688–1698.
Article15. Jézéquel P, Loussouarn D, Guérin-Charbonnel C, Campion L, Vanier A, Gouraud W, et al. Gene-expression molecular subtyping of triple-negative breast cancer tumours: importance of immune response. Breast Cancer Res. 2015; 17:43.
Article16. Ahn SG, Kim SJ, Kim C, Jeong J. Molecular classification of triple-negative breast cancer. J Breast Cancer. 2016; 19:223–230.
Article17. Kanwal R, Gupta K, Gupta S. Cancer epigenetics: an introduction. Methods Mol Biol. 2015; 1238:3–25.
Article18. Basse C, Arock M. The increasing roles of epigenetics in breast cancer: implications for pathogenicity, biomarkers, prevention and treatment. Int J Cancer. 2015; 137:2785–2794.
Article19. Tammen SA, Friso S, Choi SW. Epigenetics: the link between nature and nurture. Mol Aspects Med. 2013; 34:753–764.
Article20. Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007; 128:683–692.
Article21. Elsheikh SE, Green AR, Rakha EA, Powe DG, Ahmed RA, Collins HM, et al. Global histone modifications in breast cancer correlate with tumor phenotypes, prognostic factors, and patient outcome. Cancer Res. 2009; 69:3802–3809.
Article22. Palazzo AF, Lee ES. Non-coding RNA: what is functional and what is junk? Front Genet. 2015; 6:2.
Article23. Stirzaker C, Zotenko E, Song JZ, Qu W, Nair SS, Locke WJ, et al. Methylome sequencing in triple-negative breast cancer reveals distinct methylation clusters with prognostic value. Nat Commun. 2015; 6:5899.
Article24. Harburg GC, Hinck L. Navigating breast cancer: axon guidance molecules as breast cancer tumor suppressors and oncogenes. J Mammary Gland Biol Neoplasia. 2011; 16:257–270.
Article25. Robichaux MA, Cowan CW. Signaling mechanisms of axon guidance and early synaptogenesis. Curr Top Behav Neurosci. 2014; 16:19–48.
Article26. Braicu C, Chiorean R, Irimie A, Chira S, Tomuleasa C, Neagoe E, et al. Novel insight into triple-negative breast cancers, the emerging role of angiogenesis, and antiangiogenic therapy. Expert Rev Mol Med. 2016; 18:e18.
Article27. Branham MT, Marzese DM, Laurito SR, Gago FE, Orozco JI, Tello OM, et al. Methylation profile of triple-negative breast carcinomas. Oncogenesis. 2012; 1:e17.
Article28. Greenup R, Buchanan A, Lorizio W, Rhoads K, Chan S, Leedom T, et al. Prevalence of BRCA mutations among women with triple-negative breast cancer (TNBC) in a genetic counseling cohort. Ann Surg Oncol. 2013; 20:3254–3258.
Article29. Pop LA, Cojocneanu-Petric RM, Pileczki V, Morar-Bolba G, Irimie A, Lazar V, et al. Genetic alterations in sporadic triple negative breast cancer. Breast. 2018; 38:30–38.
Article30. Lord CJ, Ashworth A. BRCAness revisited. Nat Rev Cancer. 2016; 16:110–120.
Article31. Lips EH, Mulder L, Oonk A, van der Kolk LE, Hogervorst FB, Imholz AL, et al. Triple-negative breast cancer: BRCAness and concordance of clinical features with BRCA1-mutation carriers. Br J Cancer. 2013; 108:2172–2177.
Article32. Murria R, Palanca S, de Juan I, Alenda C, Egoavil C, Seguí FJ, et al. Immunohistochemical, genetic and epigenetic profiles of hereditary and triple negative breast cancers: relevance in personalized medicine. Am J Cancer Res. 2015; 5:2330–2343.33. Sharma P, Stecklein SR, Kimler BF, Sethi G, Petroff BK, Phillips TA, et al. The prognostic value of BRCA1 promoter methylation in early stage triple negative breast cancer. J Cancer Ther Res. 2014; 3:1–11.
Article34. Stefansson OA, Villanueva A, Vidal A, Martí L, Esteller M. BRCA1 epigenetic inactivation predicts sensitivity to platinum-based chemotherapy in breast and ovarian cancer. Epigenetics. 2012; 7:1225–1229.
Article35. Veeck J, Ropero S, Setien F, Gonzalez-Suarez E, Osorio A, Benitez J, et al. BRCA1 CpG island hypermethylation predicts sensitivity to poly(adenosine diphosphate)-ribose polymerase inhibitors. J Clin Oncol. 2010; 28:e563–e564.36. Mori H, Kubo M, Nishimura R, Osako T, Arima N, Okumura Y, et al. BRCAness as a biomarker for predicting prognosis and response to anthracycline-based adjuvant chemotherapy for patients with triple-negative breast cancer. PLoS One. 2016; 11:e0167016.
Article37. Mathe A, Wong-Brown M, Locke WJ, Stirzaker C, Braye SG, Forbes JF, et al. DNA methylation profile of triple negative breast cancer-specific genes comparing lymph node positive patients to lymph node negative patients. Sci Rep. 2016; 6:33435.
Article38. Kong B, Lv ZD, Wang Y, Jin LY, Ding L, Yang ZC. Down-regulation of BRMS1 by DNA hypermethylation and its association with metastatic progression in triple-negative breast cancer. Int J Clin Exp Pathol. 2015; 8:11076–11083.39. Kagara N, Huynh KT, Kuo C, Okano H, Sim MS, Elashoff D, et al. Epigenetic regulation of cancer stem cell genes in triple-negative breast cancer. Am J Pathol. 2012; 181:257–267.
Article40. Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012; 490:61–70.41. Holm K, Hegardt C, Staaf J, Vallon-Christersson J, Jönsson G, Olsson H, et al. Molecular subtypes of breast cancer are associated with characteristic DNA methylation patterns. Breast Cancer Res. 2010; 12:R36.
Article42. Sunami E, Shinozaki M, Sim MS, Nguyen SL, Vu AT, Giuliano AE, et al. Estrogen receptor and HER2/neu status affect epigenetic differences of tumor-related genes in primary breast tumors. Breast Cancer Res. 2008; 10:R46.
Article43. Hughes LA, Melotte V, de Schrijver J, de Maat M, Smit VT, Bovée JV, et al. The CpG island methylator phenotype: what's in a name? Cancer Res. 2013; 73:5858–5868.
Article44. Roll JD, Rivenbark AG, Jones WD, Coleman WB. DNMT3b overexpression contributes to a hypermethylator phenotype in human breast cancer cell lines. Mol Cancer. 2008; 7:15.
Article45. Sandhu R, Rivenbark AG, Coleman WB. Enhancement of chemotherapeutic efficacy in hypermethylator breast cancer cells through targeted and pharmacologic inhibition of DNMT3b. Breast Cancer Res Treat. 2012; 131:385–399.
Article46. Liu YR, Jiang YZ, Xu XE, Yu KD, Jin X, Hu X, et al. Comprehensive transcriptome analysis identifies novel molecular subtypes and subtype-specific RNAs of triple-negative breast cancer. Breast Cancer Res. 2016; 18:33.
Article47. Shen X, Xie B, Ma Z, Yu W, Wang W, Xu D, et al. Identification of novel long non-coding RNAs in triple-negative breast cancer. Oncotarget. 2015; 6:21730–21739.
Article48. Chen C, Li Z, Yang Y, Xiang T, Song W, Liu S. Microarray expression profiling of dysregulated long non-coding RNAs in triple-negative breast cancer. Cancer Biol Ther. 2015; 16:856–865.
Article49. Jadaliha M, Zong X, Malakar P, Ray T, Singh DK, Freier SM, et al. Functional and prognostic significance of long non-coding RNA MALAT1 as a metastasis driver in ER negative lymph node negative breast cancer. Oncotarget. 2016; 7:40418–40436.
Article50. Su X, Malouf GG, Chen Y, Zhang J, Yao H, Valero V, et al. Comprehensive analysis of long non-coding RNAs in human breast cancer clinical subtypes. Oncotarget. 2014; 5:9864–9876.
Article51. Liu J, Shen L, Yao J, Li Y, Wang Y, Chen H, et al. Forkhead box C1 promoter upstream transcript, a novel long non-coding RNA, regulates proliferation and migration in basal-like breast cancer. Mol Med Rep. 2014; 11:3155–3159.
Article52. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010; 464:1071–1076.
Article53. Zhuang Y, Nguyen HT, Burow ME, Zhuo Y, El-Dahr SS, Yao X, et al. Elevated expression of long intergenic non-coding RNA HOTAIR in a basal-like variant of MCF-7 breast cancer cells. Mol Carcinog. 2015; 54:1656–1667.
Article54. Wang YL, Overstreet AM, Chen MS, Wang J, Zhao HJ, Ho PC, et al. Combined inhibition of EGFR and c-ABL suppresses the growth of triple-negative breast cancer growth through inhibition of HOTAIR. Oncotarget. 2015; 6:11150–11161.
Article55. Gasparini P, Cascione L, Fassan M, Lovat F, Guler G, Balci S, et al. MicroRNA expression profiling identifies a four microRNA signature as a novel diagnostic and prognostic biomarker in triple negative breast cancers. Oncotarget. 2014; 5:1174–1184.
Article56. Fkih M'hamed I, Privat M, Ponelle F, Penault-Llorca F, Kenani A, Bignon YJ. Identification of miR-10b, miR-26a, miR-146a and miR-153 as potential triple-negative breast cancer biomarkers. Cell Oncol (Dordr). 2015; 38:433–442.57. Kumaraswamy E, Wendt KL, Augustine LA, Stecklein SR, Sibala EC, Li D, et al. BRCA1 regulation of epidermal growth factor receptor (EGFR) expression in human breast cancer cells involves microRNA-146a and is critical for its tumor suppressor function. Oncogene. 2015; 34:4333–4346.
Article58. Garcia AI, Buisson M, Bertrand P, Rimokh R, Rouleau E, Lopez BS, et al. Down-regulation of BRCA1 expression by miR-146a and miR-146b-5p in triple negative sporadic breast cancers. EMBO Mol Med. 2011; 3:279–290.
Article59. Damiano V, Brisotto G, Borgna S, di Gennaro A, Armellin M, Perin T, et al. Epigenetic silencing of miR-200c in breast cancer is associated with aggressiveness and is modulated by ZEB1. Genes Chromosomes Cancer. 2017; 56:147–158.
Article60. Humphries B, Wang Z, Oom AL, Fisher T, Tan D, Cui Y, et al. MicroRNA-200b targets protein kinase Cα and suppresses triple-negative breast cancer metastasis. Carcinogenesis. 2014; 35:2254–2263.
Article61. Tsouko E, Wang J, Frigo DE, Aydoğdu E, Williams C. miR-200a inhibits migration of triple-negative breast cancer cells through direct repression of the EPHA2 oncogene. Carcinogenesis. 2015; 36:1051–1060.
Article62. Li D, Wang H, Song H, Xu H, Zhao B, Wu C, et al. The microRNAs miR-200b-3p and miR-429-5p target the LIMK1/CFL1 pathway to inhibit growth and motility of breast cancer cells. Oncotarget. 2017; 8:85276–85289.
Article63. Mathe A, Scott RJ, Avery-Kiejda KA. miRNAs and other epigenetic changes as biomarkers in triple negative breast cancer. Int J Mol Sci. 2015; 16:28347–28376.
Article64. Xi Y, Shi J, Li W, Tanaka K, Allton KL, Richardson D, et al. Histone modification profiling in breast cancer cell lines highlights commonalities and differences among subtypes. BMC Genomics. 2018; 19:150.
Article65. Khaled WT, Choon Lee S, Stingl J, Chen X, Raza Ali H, Rueda OM, et al. BCL11A is a triple-negative breast cancer gene with critical functions in stem and progenitor cells. Nat Commun. 2015; 6:5987.
Article66. Moody RR, Lo MC, Meagher JL, Lin CC, Stevers NO, Tinsley SL, et al. Probing the interaction between the histone methyltransferase/deacetylase subunit RBBP4/7 and the transcription factor BCL11A in epigenetic complexes. J Biol Chem. 2018; 293:2125–2136.
Article67. Ocaña A, Nieto-Jiménez C, Pandiella A. BET inhibitors as novel therapeutic agents in breast cancer. Oncotarget. 2017; 8:71285–71291.
Article68. Filippakopoulos P, Knapp S. Targeting bromodomains: epigenetic readers of lysine acetylation. Nat Rev Drug Discov. 2014; 13:337–356.
Article69. Sahai V, Redig AJ, Collier KA, Eckerdt FD, Munshi HG. Targeting BET bromodomain proteins in solid tumors. Oncotarget. 2016; 7:53997–54009.
Article70. Shu S, Lin CY, He HH, Witwicki RM, Tabassum DP, Roberts JM, et al. Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer. Nature. 2016; 529:413–417.
Article71. Sahni JM, Gayle SS, Bonk KL, Vite LC, Yori JL, Webb B, et al. Bromodomain and extraterminal protein inhibition blocks growth of triple-negative breast cancers through the suppression of aurora kinases. J Biol Chem. 2016; 291:23756–23768.
Article72. Nieto-Jiménez C, Alcaraz-Sanabria A, Pérez-Peña J, Corrales-Sánchez V, Serrano-Heras G, Galán-Moya EM, et al. Targeting basal-like breast tumors with bromodomain and extraterminal domain (BET) and pololike kinase inhibitors. Oncotarget. 2017; 8:19478–19490.
Article73. Vázquez R, Riveiro ME, Astorgues-Xerri L, Odore E, Rezai K, Erba E, et al. The bromodomain inhibitor OTX015 (MK-8628) exerts anti-tumor activity in triple-negative breast cancer models as single agent and in combination with everolimus. Oncotarget. 2017; 8:7598–7613.
Article74. Gregoire JM, Fleury L, Salazar-Cardozo C, Alby F, Masson V, Arimondo PB, et al. Identification of epigenetic factors regulating the mesenchyme to epithelium transition by RNA interference screening in breast cancer cells. BMC Cancer. 2016; 16:700.
Article75. Lavigne AC, Castells M, Mermet J, Kocanova S, Dalvai M, Bystricky K. Increased macroH2A1.1 expression correlates with poor survival of triple-negative breast cancer patients. PLoS One. 2014; 9:e98930.
Article76. Healey MA, Hu R, Beck AH, Collins LC, Schnitt SJ, Tamimi RM, et al. Association of H3K9me3 and H3K27me3 repressive histone marks with breast cancer subtypes in the Nurses' Health Study. Breast Cancer Res Treat. 2014; 147:639–651.
Article77. Li G, Wang D, Ma W, An K, Liu Z, Wang X, et al. Transcriptomic and epigenetic analysis of breast cancer stem cells. Epigenomics. 2018; 10:765–783.
Article78. Mottamal M, Zheng S, Huang TL, Wang G. Histone deacetylase inhibitors in clinical studies as templates for new anticancer agents. Molecules. 2015; 20:3898–3941.
Article79. Wang ZT, Chen ZJ, Jiang GM, Wu YM, Liu T, Yi YM, et al. Histone deacetylase inhibitors suppress mutant p53 transcription via HDAC8/YY1 signals in triple negative breast cancer cells. Cell Signal. 2016; 28:506–515.
Article80. Pileczki V, Pop L, Braicu C, Budisan L, Bolba Morar G, Del C Monroig-Bosque P, et al. Double gene siRNA knockdown of mutant p53 and TNF induces apoptosis in triple-negative breast cancer cells. Onco Targets Ther. 2016; 9:6921–6933.
Article81. Braicu C, Pileczki V, Pop L, Petric RC, Chira S, Pointiere E, et al. Dual targeted therapy with p53 siRNA and epigallocatechingallate in a triple negative breast cancer cell model. PLoS One. 2015; 10:e0120936.
Article82. Braicu C, Pileczki V, Irimie A, Berindan-Neagoe I. p53siRNA therapy reduces cell proliferation, migration and induces apoptosis in triple negative breast cancer cells. Mol Cell Biochem. 2013; 381:61–68.
Article83. Tate CR, Rhodes LV, Segar HC, Driver JL, Pounder FN, Burow ME, et al. Targeting triple-negative breast cancer cells with the histone deacetylase inhibitor panobinostat. Breast Cancer Res. 2012; 14:R79.
Article84. Min A, Im SA, Kim DK, Song SH, Kim HJ, Lee KH, et al. Histone deacetylase inhibitor, suberoylanilide hydroxamic acid (SAHA), enhances anti-tumor effects of the poly (ADP-ribose) polymerase (PARP) inhibitor olaparib in triple-negative breast cancer cells. Breast Cancer Res. 2015; 17:33.
Article85. Topalian SL, Drake CG, Pardoll DM. Targeting the PD-1/B7-H1(PD-L1) pathway to activate anti-tumor immunity. Curr Opin Immunol. 2012; 24:207–212.
Article86. Terranova-Barberio M, Thomas S, Ali N, Pawlowska N, Park J, Krings G, et al. HDAC inhibition potentiates immunotherapy in triple negative breast cancer. Oncotarget. 2017; 8:114156–114172.
Article87. Schech A, Kazi A, Yu S, Shah P, Sabnis G. Histone deacetylase inhibitor entinostat inhibits tumor-initiating cells in triple-negative breast cancer cells. Mol Cancer Ther. 2015; 14:1848–1857.
Article88. Palmieri D, Lockman PR, Thomas FC, Hua E, Herring J, Hargrave E, et al. Vorinostat inhibits brain metastatic colonization in a model of triple-negative breast cancer and induces DNA double-strand breaks. Clin Cancer Res. 2009; 15:6148–6157.
Article89. Mekala JR, Naushad SM, Ponnusamy L, Arivazhagan G, Sakthiprasad V, Pal-Bhadra M. Epigenetic regulation of miR-200 as the potential strategy for the therapy against triple-negative breast cancer. Gene. 2018; 641:248–258.
Article90. Zhu Y, Bai K, Yu J, Guo M. Association between histone methyltransferase hSETD1A and prognosis in patients with triple-negative breast cancer after surgery: a retrospective study in the Chinese female population. Medicine (Baltimore). 2016; 95:e3783.91. Wu Q, Madany P, Akech J, Dobson JR, Douthwright S, Browne G, et al. The SWI/SNF ATPases are required for triple negative breast cancer cell proliferation. J Cell Physiol. 2015; 230:2683–2694.
Article92. Wu Q, Madany P, Dobson JR, Schnabl JM, Sharma S, Smith TC, et al. The BRG1 chromatin remodeling enzyme links cancer cell metabolism and proliferation. Oncotarget. 2016; 7:38270–38281.
Article93. Wu Q, Sharma S, Cui H, LeBlanc SE, Zhang H, Muthuswami R, et al. Targeting the chromatin remodeling enzyme BRG1 increases the efficacy of chemotherapy drugs in breast cancer cells. Oncotarget. 2016; 7:27158–27175.
Article94. Festuccia C, Mancini A, Colapietro A, Gravina GL, Vitale F, Marampon F, et al. The first-in-class alkylating deacetylase inhibitor molecule tinostamustine shows antitumor effects and is synergistic with radiotherapy in preclinical models of glioblastoma. J Hematol Oncol. 2018; 11:32.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Clinicopathologic Characteristics and Prognosis of Early Stage Triple Negative Breast Cancer: Comparison with Non-triple Negative Group
- Fear of Cancer Recurrence and Unmet Needs in Triple Negative Breast Cancer Survivors
- Molecular Classification of Triple-Negative Breast Cancer
- Comment on “Histomorphological Factors Predicting the Response to Neoadjuvant Chemotherapy in Triple-Negative Breast Cancerâ€
- Bilateral Triple-Negative Invasive Breast Cancer with a BRCA2 Mutation, and Glioblastoma: A Case Report and Literature Review