J Vet Sci.
2018 Sep;19(5):716-720. 10.4142/jvs.2018.19.5.716.
Evidence of two genetically different lymphotropic herpesviruses present among red deer, sambar, and milu herds in China
- Affiliations
-
- 1School of Life Sciences, Ludong University, Yantai 264025, China. zhangxingxiao@ldu.edu.cn
- 2Institute of Special Economic Animal and Plant Sciences, Chinese Academy of Agricultural Sciences, Changchun 130112, China.
- 3Shandong Provincial Key Laboratory of Quality Safety Monitoring and Risk Assessment for Animal Products, Jinan 250022, China.
- KMID: 2420942
- DOI: http://doi.org/10.4142/jvs.2018.19.5.716
Abstract
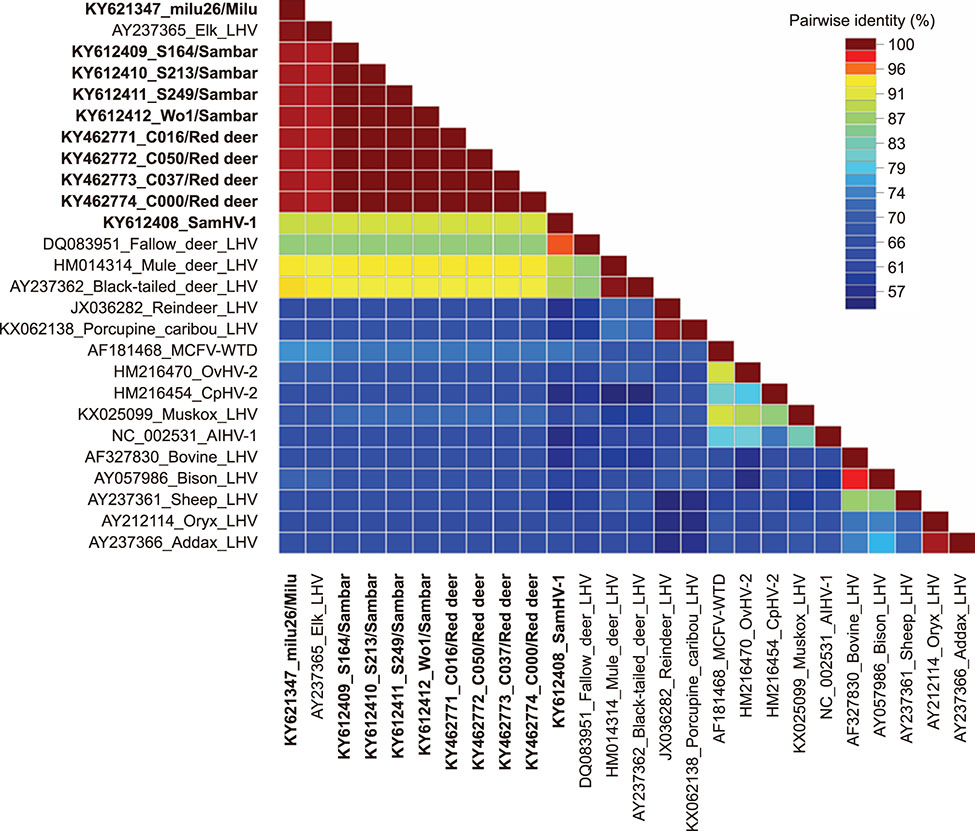
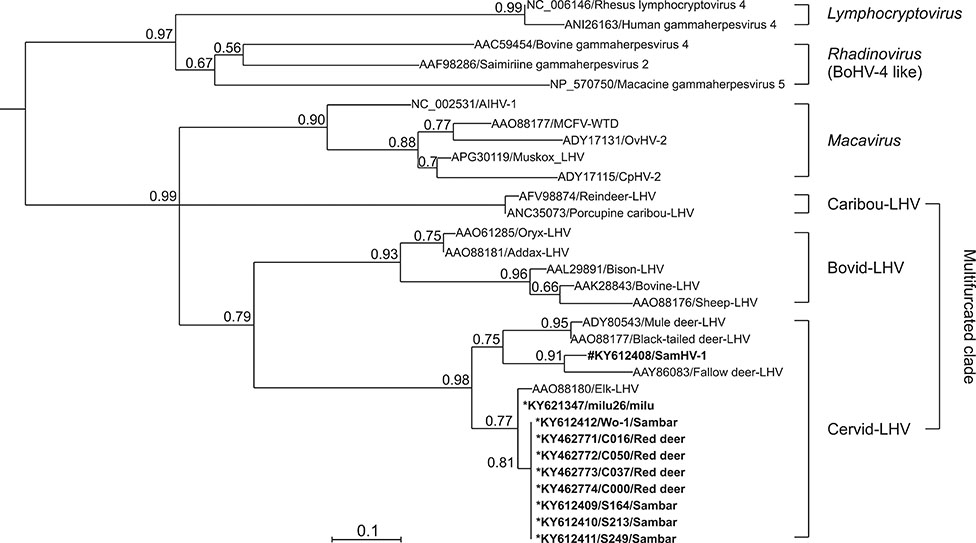
- Herpesvirus infections in Cervidae are a serious threat affecting some deer species worldwide. In our attempt to identify malignant catarrhal fever-associated herpesviruses in deer herds, ten gammaherpesviral DNA fragments were identified in five species of deer in herds in China by using a pan-herpesvirus polymerase chain reaction assay targeting viral DNA polymerase. Notably, in sambar (Rusa unicolor), a novel gamma-2 herpesvirus was identified that showed a close relationship with fallow deer lymphotropic herpesvirus (LHV), while the other fragments were phylogenetically grouped together with Elk-LHV. Determination of whether these viruses have any clinical implication in these deer species should be undertaken urgently.
MeSH Terms
Figure
Reference
-
1. Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005; 21:2104–2105.
Article2. Ackermann M. Pathogenesis of gammaherpesvirus infections. Vet Microbiol. 2006; 113:211–222.
Article3. Crawford TB, Li H, Rosenburg SR, Norhausen RW, Garner MM. Mural folliculitis and alopecia caused by infection with goat-associated malignant catarrhal fever virus in two sika deer. J Am Vet Med Assoc. 2002; 221:843–847.
Article4. Davison AJ. Evolution of the herpesviruses. Vet Microbiol. 2002; 86:69–88.
Article5. Ehlers B, Dural G, Yasmum N, Lembo T, de Thoisy B, Ryser-Degiorgis MP, Ulrich RG, McGeoch DJ. Novel mammalian herpesviruses and lineages within the Gammaherpesvirinae: cospeciation and interspecies transfer. J Virol. 2008; 82:3509–3516.
Article6. Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010; 59:307–321.
Article7. Li H, Dyer N, Keller J, Crawford TB. Newly recognized herpesvirus causing malignant catarrhal fever in white-tailed deer (Odocoileus virginianus). J Clin Microbiol. 2000; 38:1313–1318.
Article8. Li H, Gailbreath K, Flach EJ, Taus NS, Cooley J, Keller J, Russell GC, Knowles DP, Haig DM, Oaks JL, Traul DL, Crawford TB. A novel subgroup of rhadinoviruses in ruminants. J Gen Virol. 2005; 86:3021–3026.
Article9. McGeoch DJ, Rixon FJ, Davison AJ. Topics in herpesvirus genomics and evolution. Virus Res. 2006; 117:90–104.
Article10. McKillen J, Hogg K, Lagan P, Ball C, Doherty S, Reid N, Collins L, Dick JTA. Detection of a novel gammaherpesvirus (genus Rhadinovirus) in wild muntjac deer in Northern Ireland. Arch Virol. 2017; 162:1737–1740.
Article11. Moore PS, Gao SJ, Dominguez G, Cesarman E, Lungu O, Knowles DM, Garber R, Pellett PE, McGeoch DJ, Chang Y. Primary characterization of a herpesvirus agent associated with Kaposi's sarcomae. J Virol. 1996; 70:549–558.
Article12. Muhire BM, Varsani A, Martin DP. SDT: a virus classification tool based on pairwise sequence alignment and identity calculation. PLoS One. 2014; 9:e108277.
Article13. Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011; 7:539.
Article14. VanDevanter DR, Warrener P, Bennett L, Schultz ER, Coulter S, Garber RL, Rose TM. Detection and analysis of diverse herpesviral species by consensus primer PCR. J Clin Microbiol. 1996; 34:1666–1671.
Article15. Wang N, Baldi PF, Gaut BS. Phylogenetic analysis, genome evolution and the rate of gene gain in the Herpesviridae. Mol Phylogenet Evol. 2007; 43:1066–1075.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Serological and molecular epidemiology of Japanese encephalitis virus infections in swine herds in China, 2006–2012
- First Molecular Characterization of Hypoderma actaeon in Cattle and Red Deer (Cervus elaphus) in Portugal
- Seroprevalence and associated risk factors of pseudorabies in Shandong province of China
- A case of occupational asthma caused by cow Hair Cross- allergenicity between cow and deer hair allergens
- Emergence of virulent pseudorabies virus infection in Northern China