Yonsei Med J.
2017 Sep;58(5):925-933. 10.3349/ymj.2017.58.5.925.
Gene Expression Profiling of Hepatocellular Carcinoma Derived Cancer Stem Like Cell under Hypoxia
- Affiliations
-
- 1Department of Internal Medicine, Institute of Gastroenterology, Yonsei University College of Medicine, Seoul, Korea. ahnsh@yuhs.ac
- 2Division of Bioconvergence, Drug and Disease Target Group, Korea Basic Science Institute, Ochang, Korea.
- 3Department of Surgery, Yonsei University College of Medicine, Seoul, Korea.
- 4BK21 Plus Project for Medical Science, Yonsei University College of Medicine, Seoul, Korea.
- 5Yonsei Liver Center, Yonsei University Health System, Seoul, Korea.
- 6Genoplan Korea, Inc., Seoul, Korea.
- KMID: 2418927
- DOI: http://doi.org/10.3349/ymj.2017.58.5.925
Abstract
- PURPOSE
Cancer stem like cells (CSCs), with unlimited self-renewal potential and other stem cell characteristics, occur in several cancers including hepatocellular carcinoma (HCC). Although CSCs can initiate tumors, malignant proliferation, relapse and multi-drug resistance, the ways how to activate them still remain unknown. This study aims to evaluate whether CSC acquire tumorigenic characters under tumor hypoxia, analyzed by microarray analysis.
MATERIALS AND METHODS
CSCs were purified from HCC patients and Affymetrix microarray was used to investigate their gene expression profiles. The results were validated by real-time polymerase chain reaction (PCR).
RESULTS
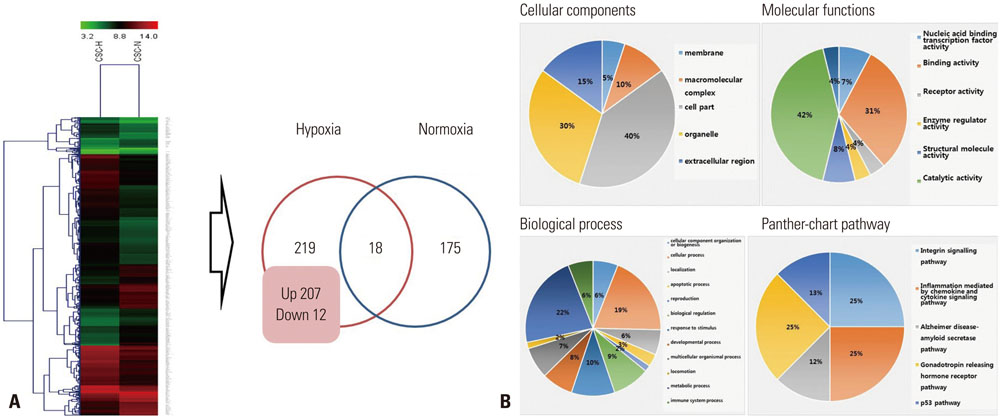
The results of the microarray indicated that 18 genes were up-regulated and 10 genes were down-regulated in CSCs. Several genes were identified to be significantly involved in the regulation of CSCs such as HCC. Furthermore, the up-regulated genes were related with metabolism, angiogenesis and hypoxia, whereas the down-regulated genes were related with apoptosis and inflammation.
CONCLUSION
The results may help to understand the mechanisms of tumor development through CSCs which acquired their distinctive tumorogenic properties by hypoxic stimulation.
MeSH Terms
-
Carcinoma, Hepatocellular/*genetics/pathology
Cell Line, Tumor
*Gene Expression Profiling
*Gene Expression Regulation, Neoplastic
Humans
Hypoxia/*complications/genetics
Liver Neoplasms/*genetics/*pathology
Neoplasm Recurrence, Local/pathology
Neoplastic Stem Cells/metabolism/*pathology
Real-Time Polymerase Chain Reaction
Reproducibility of Results
Figure
Reference
-
1. Semela D, Dufour JF. Angiogenesis and hepatocellular carcinoma. J Hepatol. 2004; 41:864–880.
Article2. Ferenci P, Fried M, Labrecque D, Bruix J, Sherman M, Omata M, et al. World Gastroenterology Organisation Guideline. Hepatocellular carcinoma (HCC): a global perspective. J Gastrointestin Liver Dis. 2010; 19:311–317.3. Harris AL. Hypoxia--a key regulatory factor in tumour growth. Nat Rev Cancer. 2002; 2:38–47.4. Samanta D, Gilkes DM, Chaturvedi P, Xiang L, Semenza GL. Hypoxia-inducible factors are required for chemotherapy resistance of breast cancer stem cells. Proc Natl Acad Sci U S A. 2014; 111:E5429–E5438.
Article5. Sun HC, Tang ZY. Angiogenesis in hepatocellular carcinoma: the retrospectives and perspectives. J Cancer Res Clin Oncol. 2004; 130:307–319.
Article6. Hickey MM, Simon MC. Regulation of angiogenesis by hypoxia and hypoxia-inducible factors. Curr Top Dev Biol. 2006; 76:217–257.
Article7. Kitajima Y, Miyazaki K. The critical impact of HIF-1a on gastric cancer biology. Cancers (Basel). 2013; 5:15–26.
Article8. Otrock ZK, Hatoum HA, Awada AH, Ishak RS, Shamseddine AI. Hypoxia-inducible factor in cancer angiogenesis: structure, regulation and clinical perspectives. Crit Rev Oncol Hematol. 2009; 70:93–102.
Article9. Pugh CW, Ratcliffe PJ. Regulation of angiogenesis by hypoxia: role of the HIF system. Nat Med. 2003; 9:677–684.
Article10. Yagi H, Kitagawa Y. The role of mesenchymal stem cells in cancer development. Front Genet. 2013; 4:261.
Article11. Krutovskikh V, Partensky C. [New insights in oncology: epigenetics and cancer stem cells]. Cancer Radiother. 2011; 15:716–722.12. Brooks DL, Schwab LP, Krutilina R, Parke DN, Sethuraman A, Hoogewijs D, et al. ITGA6 is directly regulated by hypoxia-inducible factors and enriches for cancer stem cell activity and invasion in metastatic breast cancer models. Mol Cancer. 2016; 15:26.
Article13. Heddleston JM, Li Z, Lathia JD, Bao S, Hjelmeland AB, Rich JN. Hypoxia inducible factors in cancer stem cells. Br J Cancer. 2010; 102:789–795.
Article14. Ji N, Yu JW, Ni XC, Wu JG, Wang SL, Jiang BJ. Bone marrow-derived mesenchymal stem cells increase drug resistance in CD133-expressing gastric cancer cells by regulating the PI3K/AKT pathway. Tumour Biol. 2016; 37:14637–14651.
Article15. Yeung TM, Gandhi SC, Bodmer WF. Hypoxia and lineage specification of cell line-derived colorectal cancer stem cells. Proc Natl Acad Sci U S A. 2011; 108:4382–4387.
Article16. Yamashita T, Kaneko S. Orchestration of hepatocellular carcinoma development by diverse liver cancer stem cells. J Gastroenterol. 2014; 49:1105–1110.
Article17. SchÖning JP, Monteiro M, Gu W. Drug resistance and cancer stem cells: the shared but distinct roles of hypoxia-inducible factors HIF1α and HIF2α. Clin Exp Pharmacol Physiol. 2017; 44:153–161.
Article18. Klonisch T, Wiechec E, Hombach-Klonisch S, Ande SR, Wesselborg S, Schulze-Osthoff K, et al. Cancer stem cell markers in common cancers-therapeutic implications. Trends Mol Med. 2008; 14:450–460.19. Yamashita T, Wang XW. Cancer stem cells in the development of liver cancer. J Clin Invest. 2013; 123:1911–1918.
Article20. Mathonnet M, Perraud A, Christou N, Akil H, Melin C, Battu S, et al. Hallmarks in colorectal cancer: angiogenesis and cancer stemlike cells. World J Gastroenterol. 2014; 20:4189–4196.
Article21. Pasquier J, Rafii A. Role of the microenvironment in ovarian cancer stem cell maintenance. Biomed Res Int. 2013; 2013:630782.
Article22. Bao B, Ahmad A, Li Y, Azmi AS, Ali S, Banerjee S, et al. Targeting CSCs within the tumor microenvironment for cancer therapy: a potential role of mesenchymal stem cells. Expert Opin Ther Targets. 2012; 16:1041–1054.
Article23. Bar EE. Glioblastoma, cancer stem cells and hypoxia. Brain Pathol. 2011; 21:119–129.
Article24. Gadaleta CD, Ranieri G. Trans-arterial chemoembolization as a therapy for liver tumours: new clinical developments and suggestions for combination with angiogenesis inhibitors. Crit Rev Oncol Hematol. 2011; 80:40–53.
Article25. Piao LS, Hur W, Kim TK, Hong SW, Kim SW, Choi JE, et al. CD133+ liver cancer stem cells modulate radioresistance in human hepatocellular carcinoma. Cancer Lett. 2012; 315:129–137.
Article26. Rosa R, D'Amato V, De Placido S, Bianco R. Approaches for targeting cancer stem cells drug resistance. Expert Opin Drug Discov. 2016; 11:1201–1212.
Article27. Leake I. Colorectal cancer: targeting self-renewal of cancer stem cells in colorectal cancer-a future treatment option? Nat Rev Gastroenterol Hepatol. 2014; 11:75.
Article28. Choi SH, Kwon OJ, Park JY, Kim DY, Ahn SH, Kim SU, et al. Inhibition of tumour angiogenesis and growth by small hairpin HIF-1α and IL-8 in hepatocellular carcinoma. Liver Int. 2014; 34:632–642.
Article29. Ghoshal S, Fuchs BC, Tanabe KK. STAT3 is a key transcriptional regulator of cancer stem cell marker CD133 in HCC. Hepatobiliary Surg Nutr. 2016; 5:201–203.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Translational Application of Single-cell Transcriptomic Analysis in Hepatocellular Carcinoma
- Stem Cell ; New Paradigm in the Era of Genomic Medicine
- Controlled Gene Expression System under Hypoxia Conditions
- Studies on the Mechanism of Hypoxic Increase of VEGF Expression in the Hep3B Human Hepatoma Cells
- Hypoxia in Hepatocellular Carcinoma