Cancer Res Treat.
2018 Apr;50(2):356-365. 10.4143/crt.2017.125.
Omic Approach in Non-smoker Female with Lung Squamous Cell Carcinoma Pinpoints to Germline Susceptibility and Personalized Medicine
- Affiliations
-
- 1Medical Genetics Unit, University of Siena, Siena, Italy. alessandra.renieri@unisi.it
- 2Genetica Medica, Azienda Ospedaliera Universitaria Senese, Siena, Italy.
- 3IRCCS MultiMedica, Milan, Italy.
- 4Thoracic Surgery Unit, Azienda Ospedaliera Universitaria Senese, Siena, Italy.
- 5Section of Pathology, Department of Medical Biotechnology, Chemistry and Pharmacy, University of Siena, Siena, Italy.
- 6Department of Medical Biotechnology, Chemistry and Pharmacy, University of Siena, Siena, Italy.
- 7Department of Biotechnology, Chemistry and Pharmacy, University of Siena, Siena, Italy.
- 8Medical Oncology Unit, Azienda Usl Toscana Sudest, Siena, Italy.
- KMID: 2411125
- DOI: http://doi.org/10.4143/crt.2017.125
Abstract
- PURPOSE
Lung cancer is strongly associated to tobacco smoking. However, global statistics estimate that in females the proportion of lung cancer cases that is unrelated to tobacco smoking reaches fifty percent, making questionable the etiology of the disease.
MATERIALS AND METHODS
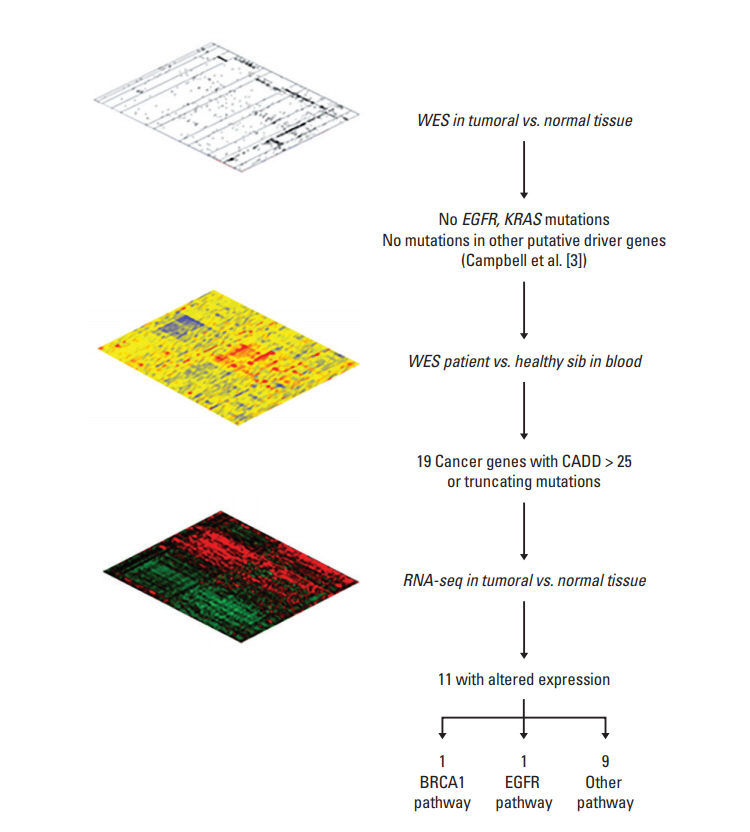
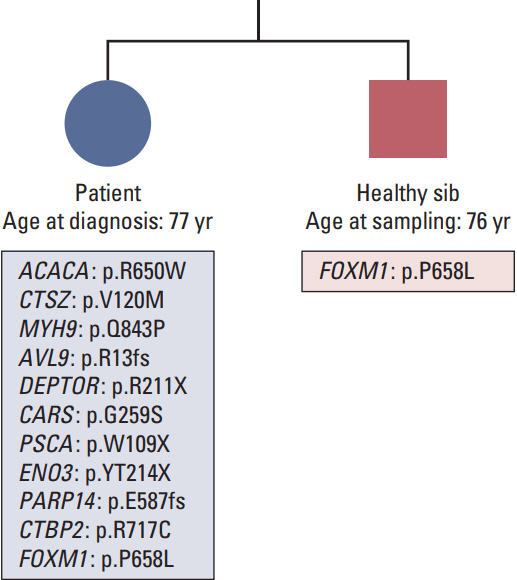
A never-smoker female with primary EGFR/KRAS/ALK-negative squamous cell carcinoma of the lung and their normal sibswere subjected to a novel integrative "omic" approach using a pedigree-based model for discovering genetic factors leading to cancer in the absence of well-known environmental trigger. A first-stepwhole-exome sequencing on tumor and normal tissue did not identify mutations in known driver genes. Building on the idea of a germline oligogenic origin of lung cancer, we performed whole-exome sequencing of DNA from patients' peripheral blood and their unaffected sibs. Finally, RNA-sequencing analysis in tumoral and matched non-tumoral tissues was carried out in order to investigate the clonal profile and the pathogenic role of the identified variants.
RESULTS
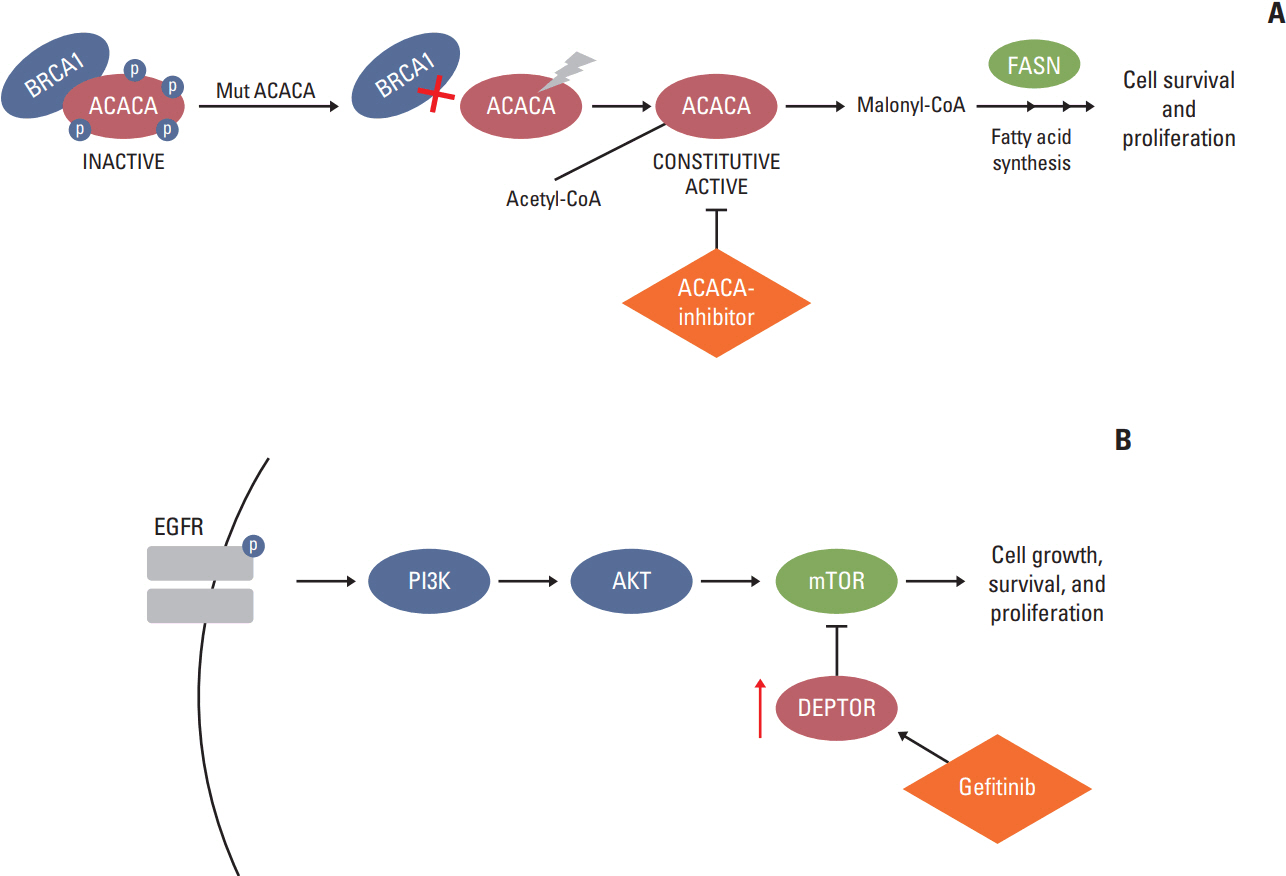
Filtering for rare variants with Combined Annotation Dependent Depletion (CADD) > 25 and potentially damaging effect, we identified rare/private germline deleterious variants in 11 cancer-associated genes, none ofwhich, except one, sharedwith the healthy sib, pinpointing to a "private" oligogenic germline signature. Noteworthy, among these, two mutated genes, namely ACACA and DEPTOR, turned to be potential targets for therapy because related to known drivers, such as BRCA1 and EGFR.
CONCLUSION
In the era of precision medicine, this report emphasizes the importance of an "omic" approach to uncover oligogenic germline signature underlying cancer development and to identify suitable therapeutic targets as well.
Keyword
MeSH Terms
Figure
Reference
-
References
1. Stewart BW, Wild CP. World cancer report 2014. Lyon: International Agency for Research on Cancer;2014.2. Samet JM, Avila-Tang E, Boffetta P, Hannan LM, Olivo-Marston S, Thun MJ, et al. Lung cancer in never smokers: clinical epidemiology and environmental risk factors. Clin Cancer Res. 2009; 15:5626–45.
Article3. Campbell JD, Alexandrov A, Kim J, Wala J, Berger AH, Pedamallu CS, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet. 2016; 48:607–16.
Article4. Vanni I, Coco S, Bonfiglio S, Cittaro D, Genova C, Biello F, et al. Whole exome sequencing of independent lung adenocarcinoma, lung squamous cell carcinoma, and malignant peritoneal mesothelioma: a case report. Medicine (Baltimore). 2016; 95:e5447.5. Renieri A, Mencarelli MA, Cetta F, Baldassarri M, Mari F, Furini S, et al. Oligogenic germline mutations identified in early non-smokers lung adenocarcinoma patients. Lung Cancer. 2014; 85:168–74.
Article6. Imperatore V, Mencarelli MA, Fallerini C, Bianciardi L, Ariani F, Furini S, et al. Potentially treatable disorder diagnosed post mortem by exome analysis in a boy with respiratory distress. Int J Mol Sci. 2016; 17:306.
Article7. Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009; 25:1105–11.
Article8. Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012; 7:562–78.
Article9. Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010; 28:511–5.
Article10. Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013; 500:415–21.11. Magnard C, Bachelier R, Vincent A, Jaquinod M, Kieffer S, Lenoir GM, et al. BRCA1 interacts with acetyl-CoA carboxylase through its tandem of BRCT domains. Oncogene. 2002; 21:6729–39.
Article12. Chiu HC, Chang TY, Huang CT, Chao YS, Hsu JT. EGFR and myosin II inhibitors cooperate to suppress EGFR-T790Mmutant NSCLC cells. Mol Oncol. 2012; 6:299–310.
Article13. Zavasnik-Bergant T, Turk B. Cysteine cathepsins in the immune response. Tissue Antigens. 2006; 67:349–55.
Article14. Tang J, Li Y, Lyon K, Camps J, Dalton S, Ried T, et al. Cancer driver-passenger distinction via sporadic human and dog cancer comparison: a proof-of-principle study with colorectal cancer. Oncogene. 2014; 33:814–22.
Article15. Linford A, Yoshimura S, Nunes Bastos R, Langemeyer L, Gerondopoulos A, Rigden DJ, et al. Rab14 and its exchange factor FAM116 link endocytic recycling and adherens junction stability in migrating cells. Dev Cell. 2012; 22:952–66.
Article16. Hu RJ, Lee MP, Connors TD, Johnson LA, Burn TC, Su K, et al. A 2.5-Mb transcript map of a tumor-suppressing subchromosomal transferable fragment from 11p15.5, and isolation and sequence analysis of three novel genes. Genomics. 1997; 46:9–17.
Article17. Li H, Sun GY, Zhao Y, Thomas D, Greenson JK, Zalupski MM, et al. DEPTOR has growth suppression activity against pancreatic cancer cells. Oncotarget. 2014; 5:12811–9.
Article18. Zhou X, Guo J, Ji Y, Pan G, Liu T, Zhu H, et al. Reciprocal negative regulation between EGFR and DEPTOR plays an important role in the progression of lung adenocarcinoma. Mol Cancer Res. 2016; 14:448–57.
Article19. Kawaguchi T, Sho M, Tojo T, Yamato I, Nomi T, Hotta K, et al. Clinical significance of prostate stem cell antigen expression in non-small cell lung cancer. Jpn J Clin Oncol. 2010; 40:319–26.
Article20. Li JL, Fei Q, Yu J, Zhang HY, Wang P, Zhu JD. Correlation between methylation profile of promoter cpg islands of seven metastasis-associated genes and their expression states in six cell lines of liver origin. Ai Zheng. 2004; 23:985–91.21. Barbarulo A, Iansante V, Chaidos A, Naresh K, Rahemtulla A, Franzoso G, et al. Poly(ADP-ribose) polymerase family member 14 (PARP14) is a novel effector of the JNK2-dependent pro-survival signal in multiple myeloma. Oncogene. 2013; 32:4231–42.
Article22. Zhang C, Li S, Qiao B, Yang K, Liu R, Ma B, et al. CtBP2 overexpression is associated with tumorigenesis and poor clinical outcome of prostate cancer. Arch Med Sci. 2015; 11:1318–23.
Article23. Wang Y, Che S, Cai G, He Y, Chen J, Xu W. Expression and prognostic significance of CTBP2 in human gliomas. Oncol Lett. 2016; 12:2429–34.
Article24. Kim IM, Ackerson T, Ramakrishna S, Tretiakova M, Wang IC, Kalin TV, et al. The Forkhead Box m1 transcription factor stimulates the proliferation of tumor cells during development of lung cancer. Cancer Res. 2006; 66:2153–61.
Article25. Tong L. Acetyl-coenzyme A carboxylase: crucial metabolic enzyme and attractive target for drug discovery. Cell Mol Life Sci. 2005; 62:1784–803.
Article26. Currie E, Schulze A, Zechner R, Walther TC, Farese RV Jr. Cellular fatty acid metabolism and cancer. Cell Metab. 2013; 18:153–61.
Article27. Moreau K, Dizin E, Ray H, Luquain C, Lefai E, Foufelle F, et al. BRCA1 affects lipid synthesis through its interaction with acetyl-CoA carboxylase. J Biol Chem. 2006; 281:3172–81.
Article28. Singh R, Yadav V, Kumar S, Saini N. MicroRNA-195 inhibits proliferation, invasion and metastasis in breast cancer cells by targeting FASN, HMGCR, ACACA and CYP27B1. Sci Rep. 2015; 5:17454.
Article29. Cordonier EL, Jarecke SK, Hollinger FE, Zempleni J. Inhibition of acetyl-CoA carboxylases by soraphen A prevents lipid accumulation and adipocyte differentiation in 3T3-L1 cells. Eur J Pharmacol. 2016; 780:202–8.
Article30. Peterson TR, Laplante M, Thoreen CC, Sancak Y, Kang SA, Kuehl WM, et al. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell. 2009; 137:873–86.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Multiple primary lung cancer: Synchronous small cell lung carcinoma and squamous cell carcinoma
- The National Survey of Lung Cancer in Korea
- Druggable Targets of Squamous Cell Lung Cancer
- Vulvar Skin Metastasis of Lung Squamous Cell Carcinoma
- Squamous Cell Carcinoma of Lung mixed with Malignant Lymphoma: 1 case report