J Pathol Transl Med.
2018 Mar;52(2):71-78. 10.4132/jptm.2017.10.21.
Protein Phosphatase Magnesium-Dependent 1δ (PPM1D) Expression as a Prognostic Marker in Adult Supratentorial Diffuse Astrocytic and Oligodendroglial Tumors
- Affiliations
-
- 1Department of Pathology, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea. jenec@amc.seoul.kr
- 2Department of Pathology, Chungbuk National University Hospital, Chungbuk National University College of Medicine, Cheongju, Korea.
- 3Department of Biostatistics, Clinical Trial Center, Soonchunhyang Medical Center, Bucheon, Korea.
- KMID: 2408506
- DOI: http://doi.org/10.4132/jptm.2017.10.21
Abstract
- BACKGROUND
Protein phosphatase magnesium-dependent 1δ (PPM1D) is a p53-induced serine/threonine phosphatase, which is overexpressed in various human cancers. A recent study reported that a mutation in the PPM1D gene is associated with poor prognosis in brainstem gliomas. In this study, we evaluated the utility of PPM1D as a prognostic biomarker of adult supratentorial diffuse astrocytic and oligodendroglial tumors.
METHODS
To investigate PPM1D protein expression, mRNA expression, and copy number changes, immunohistochemistry, RNAscope in situ hybridization, and fluorescence in situ hybridization were performed in 84 adult supratentorial diffuse gliomas. We further analyzed clinical characteristics and overall survival (OS) according to PPM1D protein expression, and examined its correlation with other glioma biomarkers such as isocitrate dehydrogenase (IDH) mutation, and p53 expression.
RESULTS
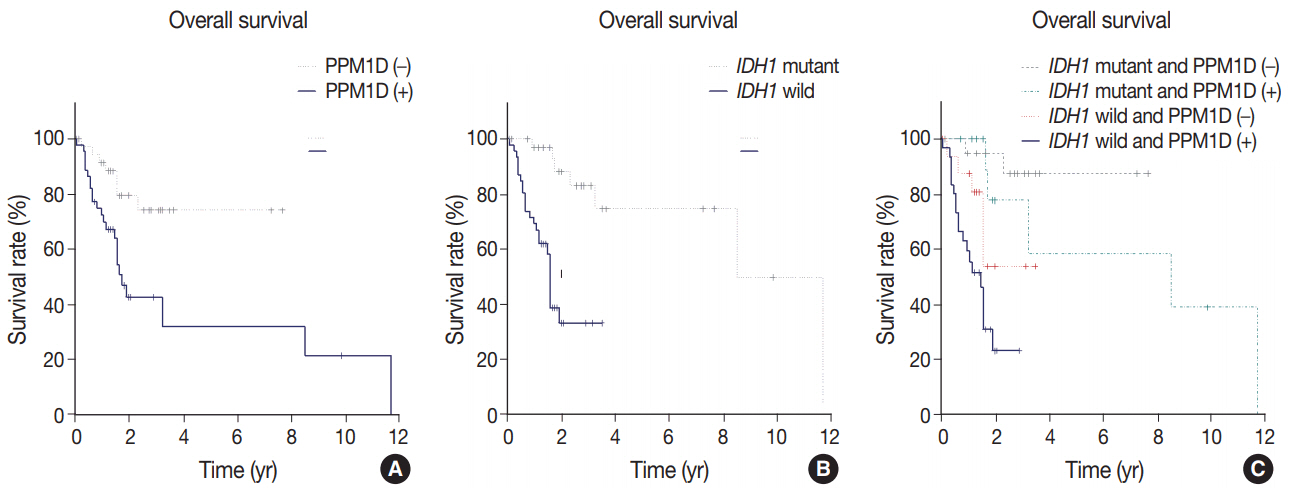
Forty-six cases (54.8%) were PPM1D-positive. PPM1D expression levels were significantly correlated with PPM1D transcript levels (p=.035), but marginally with PPM1D gene amplification (p=.079). Patients with high-grade gliomas showed a higher frequency of PPM1D expression than those with low-grade gliomas (p < .001). Multivariate analysis demonstrated that PPM1D expression (hazard ratio [HR], 2.58; p=.032), age over 60 years (HR, 2.55; p=.018), and IDH1 mutation (HR, 0.18; p=.002) were significantly independent prognostic factors; p53 expression had no prognostic significance (p=.986). The patients with tumor expressing PPM1D showed a shorter OS (p=.003). Moreover, patients with tumor harboring wild-type IDH1 and PPM1D expression had the worst OS (p < .001).
CONCLUSIONS
Our data suggest that a subset of gliomas express PPM1D; PPM1D expression is a significant marker of poor prognosis in adult supratentorial diffuse astrocytic and oligodendroglial tumors.
Keyword
MeSH Terms
Figure
Reference
-
1. Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008; 359:492–507.
Article2. DeAngelis LM. Brain tumors. N Engl J Med. 2001; 344:114–23.
Article3. Brennan CW, Verhaak RG, McKenna A, et al. The somatic genomic landscape of glioblastoma. Cell. 2013; 155:462–77.4. Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008; 455:1061–8.5. Cancer Genome Atlas Research Network, Brat DJ, Verhaak RG, et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015; 372:2481–98.
Article6. Chen R, Smith-Cohn M, Cohen AL, Colman H. Glioma subclassifications and their clinical significance. Neurotherapeutics. 2017; 14:284–97.
Article7. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016; 131:803–20.
Article8. Reuss DE, Sahm F, Schrimpf D, et al. ATRX and IDH1-R132H immunohistochemistry with subsequent copy number analysis and IDH sequencing as a basis for an "integrated" diagnostic approach for adult astrocytoma, oligodendroglioma and glioblastoma. Acta Neuropathol. 2015; 129:133–46.
Article9. Suzuki H, Aoki K, Chiba K, et al. Mutational landscape and clonal architecture in grade II and III gliomas. Nat Genet. 2015; 47:458–68.
Article10. Lass U, Numann A, von Eckardstein K, et al. Clonal analysis in recurrent astrocytic, oligoastrocytic and oligodendroglial tumors implicates IDH1-mutation as common tumor initiating event. PLoS One. 2012; 7:e41298.11. Labussière M, Idbaih A, Wang XW, et al. All the 1p19q codeleted gliomas are mutated on IDH1 or IDH2. Neurology. 2010; 74:1886–90.
Article12. Nagpal J, Jamoona A, Gulati ND, et al. Revisiting the role of p53 in primary and secondary glioblastomas. Anticancer Res. 2006; 26:4633–9.13. Tabouret E, Nguyen AT, Dehais C, et al. Prognostic impact of the 2016 WHO classification of diffuse gliomas in the French POLA cohort. Acta Neuropathol. 2016; 132:625–34.
Article14. Dudgeon C, Shreeram S, Tanoue K, et al. Genetic variants and mutations of PPM1D control the response to DNA damage. Cell Cycle. 2013; 12:2656–64.15. Lee DH, Chowdhury D. What goes on must come off: phosphatases gate-crash the DNA damage response. Trends Biochem Sci. 2011; 36:569–77.
Article16. Le Guezennec X, Bulavin DV. WIP1 phosphatase at the crossroads of cancer and aging. Trends Biochem Sci. 2010; 35:109–14.
Article17. Lu X, Nguyen TA, Moon SH, Darlington Y, Sommer M, Donehower LA. The type 2C phosphatase Wip1: an oncogenic regulator of tumor suppressor and DNA damage response pathways. Cancer Metastasis Rev. 2008; 27:123–35.
Article18. Song JY, Ryu SH, Cho YM, et al. Wip1 suppresses apoptotic cell death through direct dephosphorylation of BAX in response to gamma-radiation. Cell Death Dis. 2013; 4:e744.19. Bulavin DV, Demidov ON, Saito S, et al. Amplification of PPM1D in human tumors abrogates p53 tumor-suppressor activity. Nat Genet. 2002; 31:210–5.
Article20. Kleiblova P, Shaltiel IA, Benada J, et al. Gain-of-function mutations of PPM1D/Wip1 impair the p53-dependent G1 checkpoint. J Cell Biol. 2013; 201:511–21.
Article21. Lu X, Nannenga B, Donehower LA. PPM1D dephosphorylates Chk1 and p53 and abrogates cell cycle checkpoints. Genes Dev. 2005; 19:1162–74.
Article22. Lu X, Ma O, Nguyen TA, Jones SN, Oren M, Donehower LA. The Wip1 phosphatase acts as a gatekeeper in the p53-Mdm2 autoregulatory loop. Cancer Cell. 2007; 12:342–54.
Article23. Hirasawa A, Saito-Ohara F, Inoue J, et al. Association of 17q21-q24 gain in ovarian clear cell adenocarcinomas with poor prognosis and identification of PPM1D and APPBP2 as likely amplification targets. Clin Cancer Res. 2003; 9:1995–2004.24. Hu W, Feng Z, Modica I, et al. Gene amplifications in well-differentiated pancreatic neuroendocrine tumors inactivate the p53 pathway. Genes Cancer. 2010; 1:360–8.
Article25. Lambros MB, Natrajan R, Geyer FC, et al. PPM1D gene amplification and overexpression in breast cancer: a qRT-PCR and chromogenic in situ hybridization study. Mod Pathol. 2010; 23:1334–45.
Article26. Loukopoulos P, Shibata T, Katoh H, et al. Genome-wide array-based comparative genomic hybridization analysis of pancreatic adenocarcinoma: identification of genetic indicators that predict patient outcome. Cancer Sci. 2007; 98:392–400.
Article27. Saito-Ohara F, Imoto I, Inoue J, et al. PPM1D is a potential target for 17q gain in neuroblastoma. Cancer Res. 2003; 63:1876–83.28. Tan DS, Lambros MB, Rayter S, et al. PPM1D is a potential therapeutic target in ovarian clear cell carcinomas. Clin Cancer Res. 2009; 15:2269–80.
Article29. Yu E, Ahn YS, Jang SJ, et al. Overexpression of the wip1 gene abrogates the p38 MAPK/p53/Wip1 pathway and silences p16 expression in human breast cancers. Breast Cancer Res Treat. 2007; 101:269–78.
Article30. Liang C, Guo E, Lu S, et al. Over-expression of wild-type p53-induced phosphatase 1 confers poor prognosis of patients with gliomas. Brain Res. 2012; 1444:65–75.
Article31. Wang P, Rao J, Yang H, Zhao H, Yang L. Wip1 over-expression correlated with TP53/p14(ARF) pathway disruption in human astrocytomas. J Surg Oncol. 2011; 104:679–84.
Article32. Zhang L, Chen LH, Wan H, et al. Exome sequencing identifies somatic gain-of-function PPM1D mutations in brainstem gliomas. Nat Genet. 2014; 46:726–30.
Article33. Chuman Y, Kurihashi W, Mizukami Y, Nashimoto T, Yagi H, Sakaguchi K. PPM1D430, a novel alternative splicing variant of the human PPM1D, can dephosphorylate p53 and exhibits specific tissue expression. J Biochem. 2009; 145:1–12.
Article34. Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004; 10:7252–9.35. Wang X, Chen JX, Liu JP, You C, Liu YH, Mao Q. Gain of function of mutant TP53 in glioblastoma: prognosis and response to temozolomide. Ann Surg Oncol. 2014; 21:1337–44.
Article36. Watanabe T, Katayama Y, Yoshino A, Komine C, Yokoyama T. Deregulation of the TP53/p14ARF tumor suppressor pathway in low-grade diffuse astrocytomas and its influence on clinical course. Clin Cancer Res. 2003; 9:4884–90.37. Jones C, Baker SJ. Unique genetic and epigenetic mechanisms driving paediatric diffuse high-grade glioma. Nat Rev Cancer. 2014; 14:651–61.
Article38. Karsy M, Neil JA, Guan J, Mahan MA, Colman H, Jensen RL. A practical review of prognostic correlations of molecular biomarkers in glioblastoma. Neurosurg Focus. 2015; 38:E4.
Article39. Ludwig K, Kornblum HI. Molecular markers in glioma. J Neurooncol. 2017; 134:505–12.
Article40. Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012; 2:401–4.41. Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013; 6:pl1.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Expression of p27kip1 Protein in Astrocytic Tumors
- Comparative Evaluation of p53-protein Expression and Proliferating Indices in Astrocytic Tumors
- Bcl-2 and Bax Expression and Ki-67 Proliferative Index in Astrocytic Tumors: in Relation to Prognosis
- Expression of the DNA Repair Gene, N-Methylpurine-DNA Glycosylase in Astrocytic Tumors
- Reclassification of Mixed Oligoastrocytic Tumors Using a Genetically Integrated Diagnostic Approach