J Vet Sci.
2018 Jan;19(1):71-78. 10.4142/jvs.2018.19.1.71.
Isolation and characterization of a new porcine epidemic diarrhea virus variant that occurred in Korea in 2014
- Affiliations
-
- 1Animal and Plant Quarantine Agency, Ministry of Agriculture, Food and Rural Affairs, Gimcheon 39660, Korea. yangdk@korea.kr
- KMID: 2402697
- DOI: http://doi.org/10.4142/jvs.2018.19.1.71
Abstract
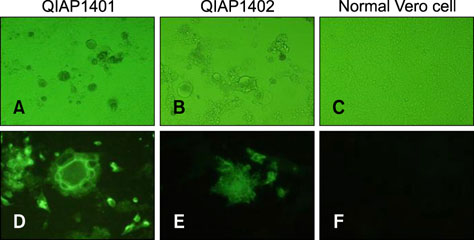
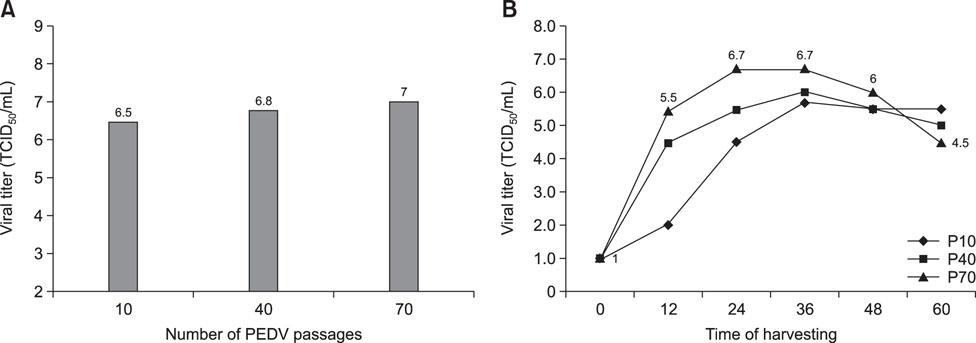
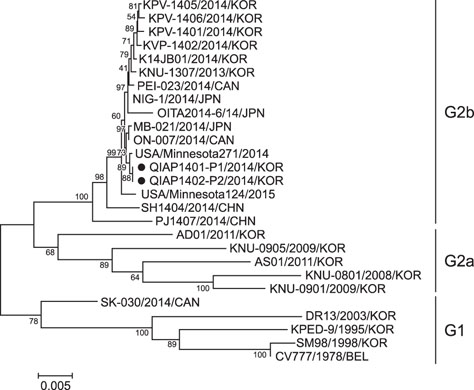
- Outbreaks of porcine epidemic diarrhea (PED) have resulted in significant economic losses in the swine industry, and another PED outbreak occurred in 2014 in Korea. Isolating and culturing PED virus (PEDV) allow investigations into its pathogenesis and the development of vaccines and diagnostic assays. In this study, we successfully isolated two PEDV isolates (QIAP1401 and QIAP1402) from naturally infected piglets at Jeju-do, Korea. Viral propagation was confirmed in Vero cells based on cytopathic effect, immunofluorescence assay, reverse transcription-polymerase chain reaction, and electron microscopic analyses. The QIAP401 isolate propagated well in Vero cells for 70 passages, with titers of 10(6.5) to 10(7.0) 50% tissue culture infectious dose/mL, which increased gradually with passaging. The nucleotide and amino acid sequences of the QIAP1401 isolate were determined and compared with those of other PEDV isolates. The QIAP1401 isolate was determined to be closely related to the USA/Minnesota271/2014 strain (> 99.9% nucleotide similarity) that was isolated in the USA in 2014. Phylogenetic analysis based on several PEDV genes suggested that a new PEDV variant is circulating in the Korean swine industry, with 93.08% similarity to the SM98 strain isolated in 1998. In addition, the QIAP1401 strain showed strong virulence in 3-day-old piglets and 11-week-old growing pigs.
Keyword
MeSH Terms
Figure
Cited by 1 articles
-
Efficacy of inactivated variant porcine epidemic diarrhea virus vaccines in growing pigs
Seung Heon Lee, Dong-Kun Yang, Ha-Hyun Kim, In-Soo Cho
Clin Exp Vaccine Res. 2018;7(1):61-69. doi: 10.7774/cevr.2018.7.1.61.
Reference
-
1. Chen Q, Gauger PC, Stafne MR, Thomas JT, Madson DM, Huang H, Zheng Y, Li G, Zhang J. Pathogenesis comparison between the United States porcine epidemic diarrhoea virus prototype and S-INDEL-variant strains in conventional neonatal piglets. J Gen Virol. 2016; 97:1107–1121.
Article2. Chen Q, Li G, Stasko J, Thomas JT, Stensland WR, Pillatzki AE, Gauger PC, Schwartz KJ, Madson D, Yoon KJ, Stevenson GW, Burrough ER, Harmon KM, Main RG, Zhang J. Isolation and characterization of porcine epidemic diarrhea viruses associated with the 2013 disease outbreak among swine in the United States. J Clin Microbiol. 2014; 52:234–243.
Article3. Choi JC, Lee KK, Pi JH, Park SY, Song CS, Choi IS, Lee JB, Lee DH, Lee SW. Comparative genome analysis and molecular epidemiology of the reemerging porcine epidemic diarrhea virus strains isolated in Korea. Infect Genet Evol. 2014; 26:348–351.
Article4. Chung HC, Nguyen VG, Moon HJ, Lee JH, Park SJ, Lee GE, Kim HK, Noh YS, Lee CH, Goede D, Park BK. Isolation of porcine epidemic diarrhea virus during outbreaks in South Korea, 2013-2014. Emerg Infect Dis. 2015; 21:2238–2240.
Article5. Crawford K, Lager K, Miller L, Opriessnig T, Gerber P, Hesse R. Evaluation of porcine epidemic diarrhea virus transmission and the immune response in growing pigs. Vet Res. 2015; 46:49.
Article6. Duarte M, Tobler K, Bridgen A, Rasschaert D, Ackermann M, Laude H. Sequence analysis of the porcine epidemic diarrhea virus genome between the nucleocapsid and spike protein genes reveals a polymorphic ORF. Virology. 1994; 198:466–476.
Article7. Fan H, Zhang J, Ye Y, Tong T, Xie K, Liao M. Complete genome sequence of a novel porcine epidemic diarrhea virus in south China. J Virol. 2012; 86:10248–10249.
Article8. Hofmann M, Wyler R. Propagation of the virus of porcine epidemic diarrhea in cell culture. J Clin Microbiol. 1988; 26:2235–2239.
Article9. Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016; 33:1870–1874.
Article10. Kusanagi K, Kuwahara H, Katoh T, Nunoya T, Ishikawa Y, Samejima T, Tajima M. Isolation and serial propagation of porcine epidemic diarrhea virus in cell cultures and partial characterization of the isolate. J Vet Med Sci. 1992; 54:313–318.
Article11. Kweon CH, Kwon BJ, Jung TS, Kee YJ, Hur DR, Hwang EK. Isolation of porcine epidemic diarrhea virus (PEDV) in Korea. Korean J Vet Res. 1993; 33:249–254.12. Lee S, Kim Y, Lee C. Isolation and characterization of a Korean porcine epidemic diarrhea virus strain KNU-141112. Virus Res. 2015; 208:215–224.
Article13. Li RF, Tian XQ, Liu Y, Xu J, Liu DY. Isolation and genetic analysis of a variant porcine epidemic diarrhea virus in China. Pol J Vet Sci. 2016; 19:65–73.
Article14. Li W, van Kuppeveld FJM, He Q, Rottier PJM, Bosch BJ. Cellular entry of the porcine epidemic diarrhea virus. Virus Res. 2016; 226:117–127.
Article15. Lin CM, Saif LJ, Marthaler D, Wang Q. Evolution, antigenicity and pathogenicity of global porcine epidemic diarrhea virus strains. Virus Res. 2016; 226:20–39.
Article16. Madson DM, Magstadt DR, Arruda PH, Hoang H, Sun D, Bower LP, Bhandari M, Burrough ER, Gauger PC, Pillatzki AE, Stevenson GW, Wilberts BL, Brodie J, Harmon KM, Wang C, Main RG, Zhang J, Yoon KJ. Pathogenesis of porcine epidemic diarrhea virus isolate (US/Iowa/18984/2013) in 3-week-old weaned pigs. Vet Microbiol. 2014; 174:60–68.
Article17. Ojkic D, Hazlett M, Fairles J, Marom A, Slavic D, Maxie G, Alexandersen S, Pasick J, Alsop J, Burlatschenko S. The first case of porcine epidemic diarrhea in Canada. Can Vet J. 2015; 56:149–152.18. Park S, Kim S, Song D, Park B. Novel porcine epidemic diarrhea virus variant with large genomic deletion, South Korea. Emerg Infect Dis. 2014; 20:2089–2092.
Article19. Pensaert MB, de Bouck P. A new coronavirus-like particle associated with diarrhea in swine. Arch Virol. 1978; 58:243–247.
Article20. Saif LJ, Pensaert MB, Sestack K, Yeo SG, Jung K. Coronaviruses. In : Straw BE, Zimmerman JJ, Karriker LA, Ramirez A, Schwartz KJ, Stevenson GW, editors. Diseases of Swine. Ames: Wiley-Blackwell;2012. p. 501–524.21. Song D, Moon H, Kang B. Porcine epidemic diarrhea: a review of current epidemiology and available vaccines. Clin Exp Vaccine Res. 2015; 4:166–176.
Article22. Song D, Park B. Porcine epidemic diarrhoea virus: a comprehensive review of molecular epidemiology, diagnosis, and vaccines. Virus Genes. 2012; 44:167–175.
Article23. Utiger A, Tobler K, Bridgen A, Ackermann M. Identification of the membrane protein of porcine epidemic diarrhea virus. Virus Genes. 1995; 10:137–148.
Article24. Zhang X, Pan Y, Wang D, Tian X, Song Y, Cao Y. Identification and pathogenicity of a variant porcine epidemic diarrhea virus field strain with reduced virulence. Virol J. 2015; 12:88.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Sequence analysis of the spike gene of Porcine epidemic diarrhea virus isolated from South China during 2011–2015
- Reemergence of porcine epidemic diarrhea virus on Jeju Island
- Genetic diversity of nucleocapsid genes of recent porcine epidemic diarrhea viruses isolated in Korea
- Recent vaccine technology in industrial animals
- Porcine epidemic diarrhea: a review of current epidemiology and available vaccines