J Korean Med Sci.
2017 May;32(5):817-824. 10.3346/jkms.2017.32.5.817.
RNA-Seq for Gene Expression Profiling of Human Necrotizing Enterocolitis: a Pilot Study
- Affiliations
-
- 1Department of Surgery, Jeju National University Hospital, Jeju, Korea.
- 2Department of Physiology, College of Medicine, Hanyang University, Seoul, Korea.
- 3Research Institute for Basic Science, Sogang University, Seoul, Korea. hdshin@sogang.ac.kr
- 4Department of Genetic Epidemiology, SNP Genetics, Inc., Seoul, Korea.
- 5Department of Surgery, Gyeongsang National University Hospital, Jinju, Korea.
- 6Department of Surgery, Donga University Hospital, Busan, Korea.
- 7Department of Surgery, Konkuk University Medical Center, Konkuk University School of Medicine, Seoul, Korea.
- 8Department of Surgery, Seoul National University Bundang Hospital, Seongnam, Korea.
- 9Department of Neonatology, Seoul National University Bundang Hospital, Seongnam, Korea.
- 10Department of Pathology, Seoul National University Bundang Hospital, Seongnam, Korea.
- 11Department of Pediatric Surgery, Seoul National University Children's Hospital, Seoul, Korea.
- 12Department of Surgery, Gyemyoung University Dongsan Hospital, Daegu, Korea.
- 13Department of Life Science, Sogang University, Seoul, Korea.
- KMID: 2375083
- DOI: http://doi.org/10.3346/jkms.2017.32.5.817
Abstract
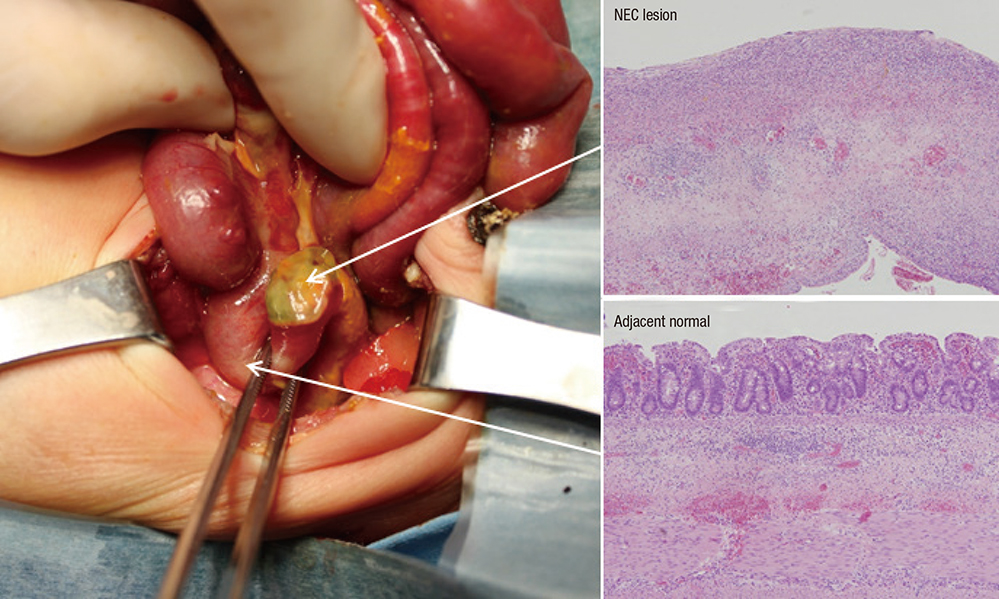
- Necrotizing enterocolitis (NEC) characterized by inflammatory intestinal necrosis is a major cause of mortality and morbidity in newborns. Deep RNA sequencing (RNA-Seq) has recently emerged as a powerful technology enabling better quantification of gene expression than microarrays with a lower background signal. A total of 10 transcriptomes from 5 pairs of NEC lesions and adjacent normal tissues obtained from preterm infants with NEC were analyzed. As a result, a total of 65 genes (57 down-regulated and 8 up-regulated) revealed significantly different expression levels in the NEC lesion compared to the adjacent normal region, based on a significance at fold change ≥ 1.5 and P ≤ 0.05. The most significant gene, DPF3 (P < 0.001), has recently been reported to have differential expressions in colon segments. Our gene ontology analysis between NEC lesion and adjacent normal tissues showed that down-regulated genes were included in nervous system development with the most significance (P = 9.3 × 10â»â·; P(corr) = 0.0003). In further pathway analysis using Pathway Express based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database, genes involved in thyroid cancer and axon guidance were predicted to be associated with different expression (P(corr) = 0.008 and 0.020, respectively). Although further replications using a larger sample size and functional evaluations are needed, our results suggest that altered gene expression and the genes' involved functional pathways and categories may provide insight into NEC development and aid in future research.
Keyword
MeSH Terms
Figure
Reference
-
1. Bisquera JA, Cooper TR, Berseth CL. Impact of necrotizing enterocolitis on length of stay and hospital charges in very low birth weight infants. Pediatrics. 2002; 109:423–428.2. Blakey JL, Lubitz L, Campbell NT, Gillam GL, Bishop RF, Barnes GL. Enteric colonization in sporadic neonatal necrotizing enterocolitis. J Pediatr Gastroenterol Nutr. 1985; 4:591–595.3. Hintz SR, Kendrick DE, Stoll BJ, Vohr BR, Fanaroff AA, Donovan EF, Poole WK, Blakely ML, Wright L, Higgins R; NICHD Neonatal Research Network. Neurodevelopmental and growth outcomes of extremely low birth weight infants after necrotizing enterocolitis. Pediatrics. 2005; 115:696–703.4. Holman RC, Stoll BJ, Clarke MJ, Glass RI. The epidemiology of necrotizing enterocolitis infant mortality in the United States. Am J Public Health. 1997; 87:2026–2031.5. Lemons JA, Bauer CR, Oh W, Korones SB, Papile LA, Stoll BJ, Verter J, Temprosa M, Wright LL, Ehrenkranz RA, et al. Very low birth weight outcomes of the National Institute of Child health and human development neonatal research network, January 1995 through December 1996. NICHD Neonatal Research Network. Pediatrics. 2001; 107:E1.6. Hunter CJ, Upperman JS, Ford HR, Camerini V. Understanding the susceptibility of the premature infant to necrotizing enterocolitis (NEC). Pediatr Res. 2008; 63:117–123.7. Morowitz MJ, Poroyko V, Caplan M, Alverdy J, Liu DC. Redefining the role of intestinal microbes in the pathogenesis of necrotizing enterocolitis. Pediatrics. 2010; 125:777–785.8. Neu J, Mihatsch W. Recent developments in necrotizing enterocolitis. JPEN J Parenter Enteral Nutr. 2012; 36:30S–35S.9. Sharma R, Hudak ML. A clinical perspective of necrotizing enterocolitis: past, present, and future. Clin Perinatol. 2013; 40:27–51.10. Sim K, Shaw AG, Randell P, Cox MJ, McClure ZE, Li MS, Haddad M, Langford PR, Cookson WO, Moffatt MF, et al. Dysbiosis anticipating necrotizing enterocolitis in very premature infants. Clin Infect Dis. 2015; 60:389–397.11. Cortese R, Lu L, Yu Y, Ruden D, Claud EC. Epigenome-Microbiome crosstalk: a potential new paradigm influencing neonatal susceptibility to disease. Epigenetics. 2016; 11:205–215.12. Christensen RD, Lambert DK, Baer VL, Gordon PV. Necrotizing enterocolitis in term infants. Clin Perinatol. 2013; 40:69–78.13. Stoll BJ. Epidemiology of necrotizing enterocolitis. Clin Perinatol. 1994; 21:205–218.14. Kanto WP Jr, Wilson R, Breart GL, Zierler S, Purohit DM, Peckham GJ, Ellison RC. Perinatal events and necrotizing enterocolitis in premature infants. Am J Dis Child. 1987; 141:167–169.15. Chan KY, Leung KT, Tam YH, Lam HS, Cheung HM, Ma TP, Lee KH, To KF, Li K, Ng PC. Genome-wide expression profiles of necrotizing enterocolitis versus spontaneous intestinal perforation in human intestinal tissues: dysregulation of functional pathways. Ann Surg. 2014; 260:1128–1137.16. Ng PC, Chan KY, Poon TC. Biomarkers for prediction and diagnosis of necrotizing enterocolitis. Clin Perinatol. 2013; 40:149–159.17. Tian R, Liu SX, Williams C, Soltau TD, Dimmitt R, Zheng X, De Plaen IG. Characterization of a necrotizing enterocolitis model in newborn mice. Int J Clin Exp Med. 2010; 3:293–302.18. Hackam DG, Redelmeier DA. Translation of research evidence from animals to humans. JAMA. 2006; 296:1731–1732.19. van der Worp HB, Howells DW, Sena ES, Porritt MJ, Rewell S, O’Collins V, Macleod MR. Can animal models of disease reliably inform human studies? PLoS Med. 2010; 7:e1000245.20. Ioannidis JP, Allison DB, Ball CA, Coulibaly I, Cui X, Culhane AC, Falchi M, Furlanello C, Game L, Jurman G, et al. Repeatability of published microarray gene expression analyses. Nat Genet. 2009; 41:149–155.21. Conesa A, Madrigal P, Tarazona S, Gomez-Cabrero D, Cervera A, McPherson A, Szcześniak MW, Gaffney DJ, Elo LL, Zhang X, et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016; 17:13.22. Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009; 10:57–63.23. Lin PW, Stoll BJ. Necrotising enterocolitis. Lancet. 2006; 368:1271–1283.24. Lucas A, Cole TJ. Breast milk and neonatal necrotising enterocolitis. Lancet. 1990; 336:1519–1523.25. Wang Y, Hoenig JD, Malin KJ, Qamar S, Petrof EO, Sun J, Antonopoulos DA, Chang EB, Claud EC. 16S rRNA gene-based analysis of fecal microbiota from preterm infants with and without necrotizing enterocolitis. ISME J. 2009; 3:944–954.26. Repa A, Thanhaeuser M, Endress D, Weber M, Kreissl A, Binder C, Berger A, Haiden N. Probiotics (Lactobacillus acidophilus and Bifidobacterium infantis) prevent NEC in VLBW infants fed breast milk but not formula. [corrected]. Pediatr Res. 2015; 77:381–388.27. Claud EC. Neonatal necrotizing enterocolitis -inflammation and intestinal immaturity. Antiinflamm Antiallergy Agents Med Chem. 2009; 8:248–259.28. Lange M, Kaynak B, Forster UB, Tönjes M, Fischer JJ, Grimm C, Schlesinger J, Just S, Dunkel I, Krueger T, et al. Regulation of muscle development by DPF3, a novel histone acetylation and methylation reader of the BAF chromatin remodeling complex. Genes Dev. 2008; 22:2370–2384.29. Liu H, Luo Y, Li S, Wang S, Wang N, Jin X. Expression profiles of HA117 and its neighboring gene DPF3 in different colon segments of Hirschsprung’s disease. Int J Clin Exp Pathol. 2014; 7:3966–3974.30. You J, Peng W, Lin X, Huang QL, Lin JY. PLC/CAMK IV-NF-kappaB involved in the receptor for advanced glycation end products mediated signaling pathway in human endothelial cells. Mol Cell Endocrinol. 2010; 320:111–117.31. Tremblay É, Thibault MP, Ferretti E, Babakissa C, Bertelle V, Bettolli M, Burghardt KM, Colombani JF, Grynspan D, Levy E, et al. Gene expression profiling in necrotizing enterocolitis reveals pathways common to those reported in Crohn’s disease. BMC Med Genomics. 2016; 9:6.32. Draghici S, Khatri P, Tarca AL, Amin K, Done A, Voichita C, Georgescu C, Romero R. A systems biology approach for pathway level analysis. Genome Res. 2007; 17:1537–1545.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- TRAPR: R Package for Statistical Analysis and Visualization of RNA-Seq Data
- Analysis of Whole Transcriptome Sequencing Data: Workflow and Software
- COEX-Seq: Convert a Variety of Measurements of Gene Expression in RNA-Seq
- Unraveling flavivirus pathogenesis: from bulk to single-cell RNA-sequencing strategies
- Blood Transcriptome Profiling in Myasthenia Gravis Patients to Assess Disease Activity: A Pilot RNA-seq Study