Restor Dent Endod.
2016 Nov;41(4):283-295. 10.5395/rde.2016.41.4.283.
In vitro characterization of human dental pulp stem cells isolated by three different methods
- Affiliations
-
- 1Department of Conservative Dentistry, Kyung Hee University Dental Hospital at Gangdong, Seoul, Korea. shpark94@khu.ac.kr
- 2Department of Pharmacology, School of Dentistry, Kyung Hee University, Seoul, Korea.
- 3Oral Biology Research Institute, School of Dentistry, Kyung Hee University, Seoul, Korea.
- 4Department of Conservative Dentistry, Graduate School, Kyung Hee University, Seoul, Korea.
- 5School of Dentistry, University of Western Australia, Nedlands, WA, Australia.
- 6School of Dentistry and Jonsson Comprehensive Cancer Center, UCLA, Los Angeles, CA, USA.
- 7Microscope Center, Department of Conservative Dentistry and Oral Science Research Center, College of Dentistry, Yonsei University, Seoul, Korea. andyendo@yuhs.ac
- KMID: 2356005
- DOI: http://doi.org/10.5395/rde.2016.41.4.283
Abstract
OBJECTIVES
In this study, we characterized human dental pulp cells (HDPCs) obtained by different culture methods to establish the most suitable methodology for dental tissue engineering and regenerative endodontic applications.
MATERIALS AND METHODS
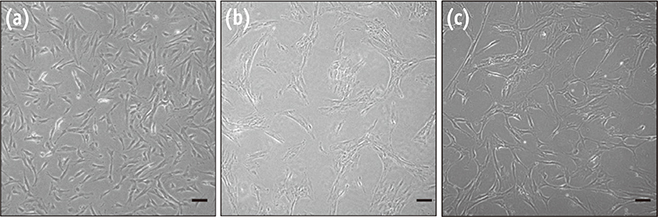
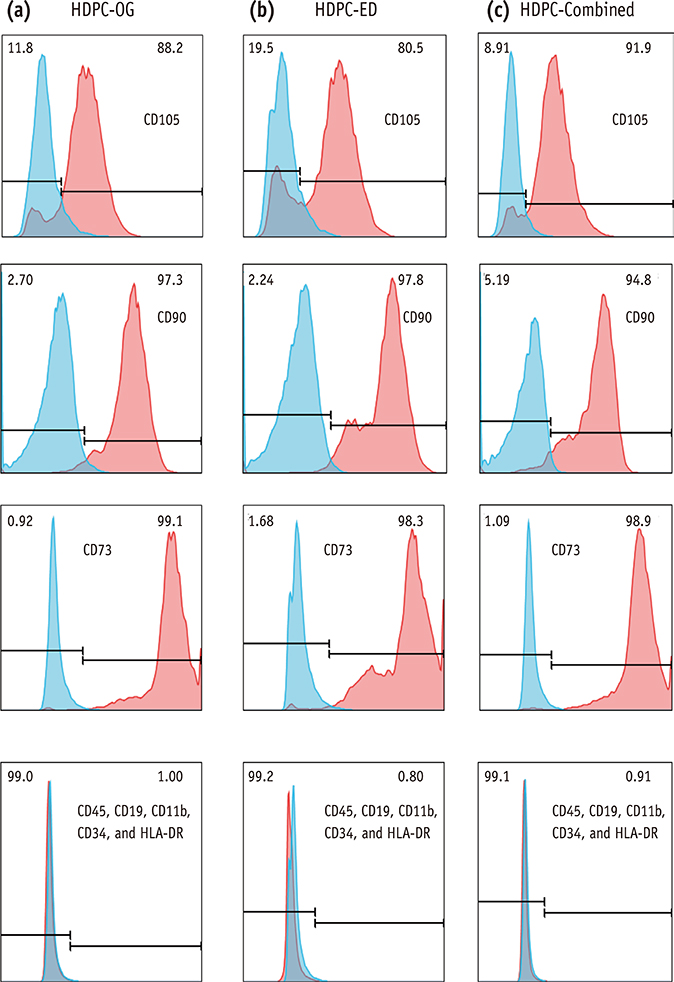
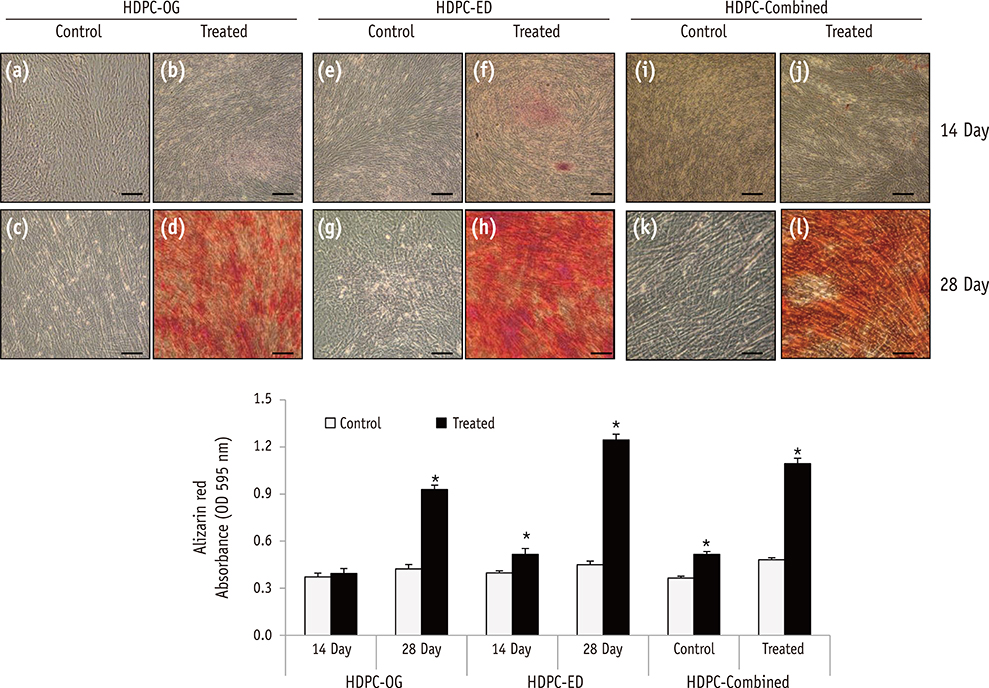
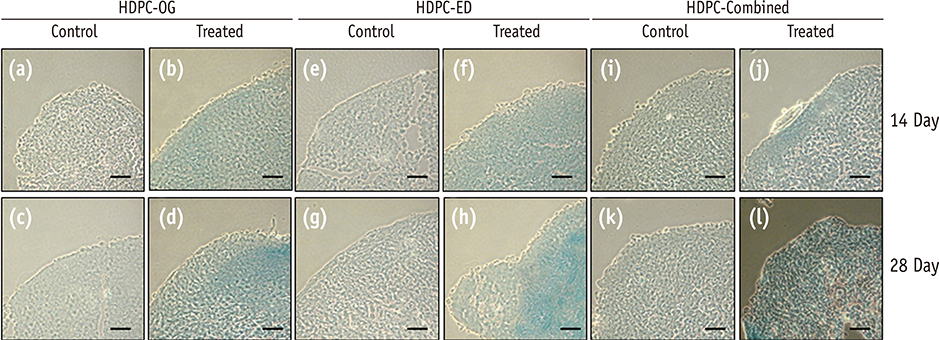
HDPCs were isolated by the outgrowth method (HDPCs-OG), the enzymatic digestion method (collagenase/dispase/trypsin, HDPCs-ED), or the combination of both methods (HDPCs-Combined). The expression of mesenchymal stem cell markers (CD105, CD90, and CD73) was investigated. In vitro differentiation capacities of HDPCs into adipogenic, osteogenic, and chondrogenic lineages were compared. Differentiation markers were analyzed by quantitative reverse-transcription polymerase chain reaction (RT-PCR) and western blotting.
RESULTS
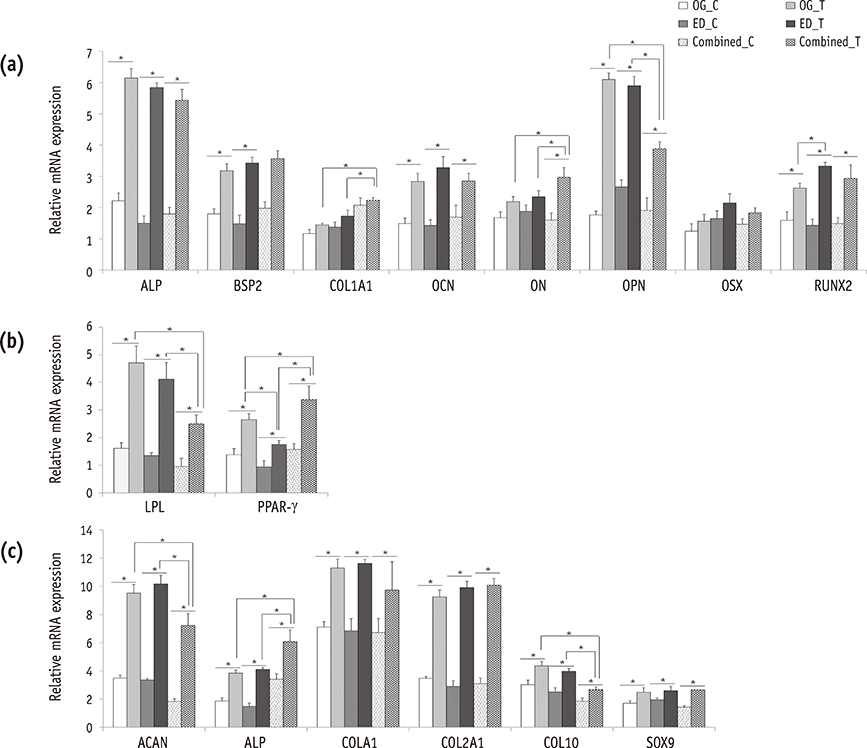
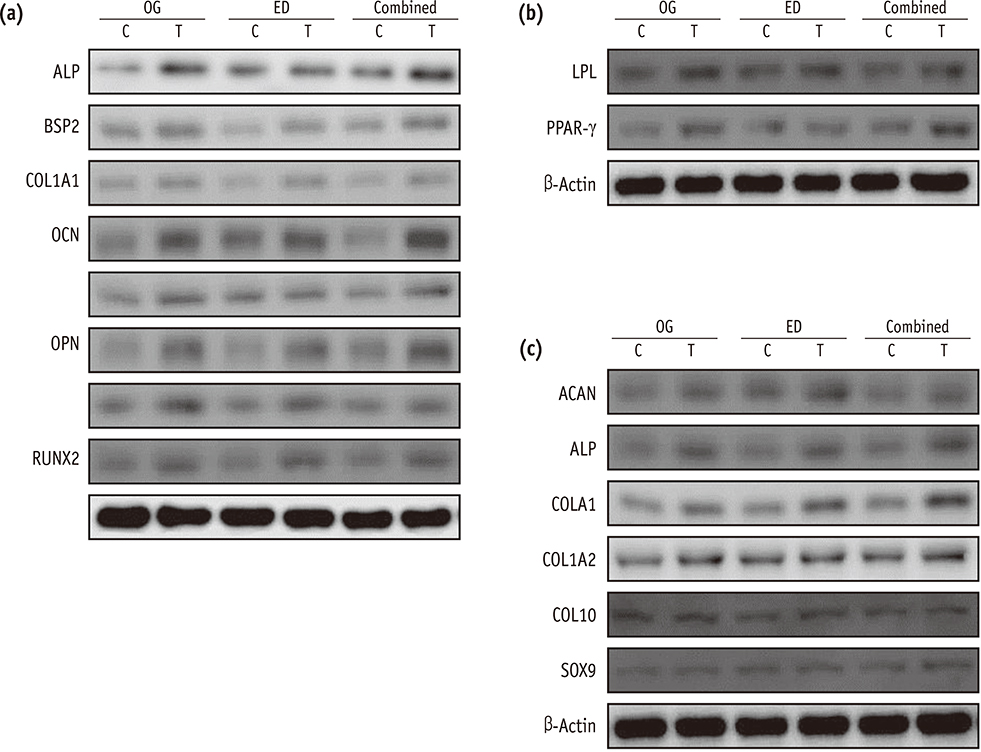
Our data indicated that whole HDPCs-ED, HPDCs-OG, and HDPCs-Combined could be differentiated into adipogenic, chrondrogenic, and osteogenic cell types. However, we found that the methods for isolating and culturing HDPCs influence the differentiation capacities of cells. HDPCs-OG and HDPCs-ED were preferably differentiated into adipogenic and osteogenic cells, respectively. Differentiation markers shown by RT-PCR and western blotting analysis were mostly upregulated in the treated groups compared with the control groups.
CONCLUSIONS
Our findings confirmed that cell populations formed by two different culture methods and the combined culture method exhibited different properties. The results of this study could provide an insight into regenerative endodontic treatment using HDPCs.
MeSH Terms
Figure
Reference
-
1. Miura M, Gronthos S, Zhao M, Lu B, Fisher LW, Robey PG, Shi S. SHED: stem cells from human exfoliated deciduous teeth. Proc Natl Acad Sci USA. 2003; 100:5807–5812.
Article2. Malhotra N, Mala K. Regenerative endodontics as a tissue engineering approach: past, current and future. Aust Endod J. 2012; 38:137–148.
Article3. About I, Bottero MJ, de Denato P, Camps J, Franquin JC, Mitsiadis TA. Human dentin production in vitro. Exp Cell Res. 2000; 258:33–41.4. Huang GT, Gronthos S, Shi S. Mesenchymal stem cells derived from dental tissues vs. those from other sources: their biology and role in regenerative medicine. J Dent Res. 2009; 88:792–806.
Article5. Gronthos S, Mankani M, Brahim J, Robey PG, Shi S. Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proc Natl Acad Sci USA. 2000; 97:13625–13630.6. Erices A, Conget P, Minguell JJ. Mesenchymal progenitor cells in human umbilical cord blood. Br J Haematol. 2000; 109:235–242.
Article7. Zuk PA, Zhu M, Mizuno H, Huang J, Futrell JW, Katz AJ, Benhaim P, Lorenz HP, Hedrick MH. Multilineage cells from human adipose tissue: implications for cell-based therapies. Tissue Eng. 2001; 7:211–228.
Article8. Mitchell KE, Weiss ML, Mitchell BM, Martin P, Davis D, Morales L, Helwig B, Beerenstrauch M, Abou-Easa K, Hildreth T, Troyer D, Medicetty S. Matrix cells from Wharton's jelly form neurons and glia. Stem Cells. 2003; 21:50–60.
Article9. Gronthos S, Brahim J, Li W, Fisher LW, Cherman N, Boyde A, DenBesten P, Robey PG, Shi S. Stem cell properties of human dental pulp stem cells. J Dent Res. 2002; 81:531–535.
Article10. Struys T, Moreels M, Martens W, Donders R, Wolfs E, Lambrichts I. Ultrastructural and immunocytochemical analysis of multilineage differentiated human dental pulp- and umbilical cord-derived mesenchymal stem cells. Cells Tissues Organs. 2011; 193:366–378.
Article11. Huang GT, Yamaza T, Shea LD, Djouad F, Kuhn NZ, Tuan RS, Shi S. Stem/progenitor cell-mediated de novo regeneration of dental pulp with newly deposited continuous layer of dentin in an in vivo model. Tissue Eng Part A. 2010; 16:605–615.
Article12. Zheng Y, Wang XY, Wang YM, Liu XY, Zhang CM, Hou BX, Wang SL. Dentin regeneration using deciduous pulp stem/progenitor cells. J Dent Res. 2012; 91:676–682.
Article13. Yang H, Shin S, Ahn J, Choi Y, Kim KH, Chung CJ. Local injection of pulp cells enhances wound healing during the initial proliferative phase through the stimulation of host angiogenesis. J Endod. 2013; 39:788–794.
Article14. Balic A, Mina M. Characterization of progenitor cells in pulps of murine incisors. J Dent Res. 2010; 89:1287–1292.
Article15. Pierdomenico L, Bonsi L, Calvitti M, Rondelli D, Arpinati M, Chirumbolo G, Becchetti E, Marchionni C, Alviano F, Fossati V, Staffolani N, Franchina M, Grossi A, Bagnara GP. Multipotent mesenchymal stem cells with immunosuppressive activity can be easily isolated from dental pulp. Transplantation. 2005; 80:836–842.
Article16. Huang GT, Sonoyama W, Chen J, Park SH. In vitro characterization of human dental pulp cells: various isolation methods and culturing environments. Cell Tissue Res. 2006; 324:225–236.
Article17. Noȅl D, Djouad F, Bouffi C, Mrugala D, Jorgensen C. Multipotent mesenchymal stromal cells and immune tolerance. Leuk Lymphoma. 2007; 48:1283–1289.
Article18. Arora V, Arora P, Munshi AK. Banking stem cells from human exfoliated deciduous teeth (SHED): saving for the future. J Clin Pediatr Dent. 2009; 33:289–294.
Article19. Sloan AJ, Waddington RJ. Dental pulp stem cells: what, where, how? Int J Paediatr Dent. 2009; 19:61–70.
Article20. Tsukamoto Y, Fukutani S, Shin-Ike T, Kubota T, Sato S, Suzuki Y, Mori M. Mineralized nodule formation by cultures of human dental pulp-derived fibroblasts. Arch Oral Biol. 1992; 37:1045–1055.
Article21. Couble ML, Farges JC, Bleicher F, Perrat-Mabillon B, Boudeulle M, Magloire H. Odontoblast differentiation of human dental pulp cells in explant cultures. Calcif Tissue Int. 2000; 66:129–138.
Article22. Onishi T, Kinoshita S, Shintani S, Sobue S, Ooshima T. Stimulation of proliferation and differentiation of dog dental pulp cells in serum-free culture medium by insulin-like growth factor. Arch Oral Biol. 1999; 44:361–371.
Article23. Bakopoulou A, Leyhausen G, Volk J, Tsiftsoglou A, Garefis P, Koidis P, Geurtsen W. Assessment of the impact of two different isolation methods on the osteo/odontogenic differentiation potential of human dental stem cells derived from deciduous teeth. Calcif Tissue Int. 2011; 88:130–141.
Article24. Park SH, Hsiao GY, Huang GT. Role of substance P and calcitonin gene-related peptide in the regulation of interleukin-8 and monocyte chemotactic protein-1 expression in human dental pulp. Int Endod J. 2004; 37:185–192.
Article25. Seo BM, Miura M, Gronthos S, Bartold PM, Batouli S, Brahim J, Young M, Robey PG, Wang CY, Shi S. Investigation of multipotent postnatal stem cells from human periodontal ligament. Lancet. 2004; 364:149–155.
Article26. Nakashima M, Nagasawa H, Yamada Y, Reddi AH. Regulatory role of transforming growth factor-beta, bone morphogenetic protein-2, and protein-4 on gene expression of extracellular matrix proteins and differentiation of dental pulp cells. Dev Biol. 1994; 162:18–28.
Article27. Sonoyama W, Liu Y, Fang D, Yamaza T, Seo BM, Zhang C, Liu H, Gronthos S, Wang CY, Wang S, Shi S. Mesenchymal stem cell-mediated functional tooth regeneration in swine. PLoS One. 2006; 1:e79.
Article28. Ellerström C, Hyllner J, Strehl R. Single cell enzymatic dissociation of human embryonic stem cells: a straightforward, robust, and standardized culture method. Methods Mol Biol. 2010; 584:121–134.
Article29. Karamzadeh R, Eslaminejad MB, Aflatoonian R. Isolation, characterization and comparative differentiation of human dental pulp stem cells derived from permanent teeth by using two different methods. J Vis Exp. 2012; (69):4372.
Article30. Qin C, Brunn JC, Cadena E, Ridall A, Tsujigiwa H, Nagatsuka H, Nagai N, Butler WT. The expression of dentin sialophosphoprotein gene in bone. J Dent Res. 2002; 81:392–394.
Article31. Viereck V, Siggelkow H, Tauber S, Raddatz D, Schutze N, Hüfner M. Differential regulation of Cbfa1/Runx2 and osteocalcin gene expression by vitamin-D3, dexamethasone, and local growth factors in primary human osteoblasts. J Cell Biochem. 2002; 86:348–356.
Article32. Fisher LW, Fedarko NS. Six genes expressed in bones and teeth encode the current members of the SIBLING family of proteins. Connect Tissue Res. 2003; 44:Suppl 1. 33–40.
Article33. Laino G, d'Aquino R, Graziano A, Lanza V, Carinci F, Naro F, Pirozzi G, Papaccio G. A new population of human adult dental pulp stem cells: a useful source of living autologous fibrous bone tissue (LAB). J Bone Miner Res. 2005; 20:1394–1402.
Article34. Laino G, Graziano A, d'Aquino R, Pirozzi G, Lanza V, Valiante S, De Rosa A, Naro F, Vivarelli E, Papaccio G. An approachable human adult stem cell source for hardtissue engineering. J Cell Physiol. 2006; 206:693–701.
Article35. Comoli P, Ginevri F, Maccario R, Avanzini MA, Marconi M, Groff A, Cometa A, Cioni M, Porretti L, Barberi W, Frassoni F, Locatelli F. Human mesenchymal stem cells inhibit antibody production induced in vitro by allostimulation. Nephrol Dial Transplant. 2008; 23:1196–1202.
Article36. Chung CR, Kim HN, Park Y, Kim MJ, Oh YJ, Shin SJ, Choi YJ, Kim KH. Morphological evaluation during in vitro chondrogenesis of dental pulp stromal cells. Restor Dent Endod. 2012; 37:34–40.
Article37. Suzuki T, Lee CH, Chen M, Zhao W, Fu SY, Qi JJ, Chotkowski G, Eisig SB, Wong A, Mao JJ. Induced migration of dental pulp stem cells for in vivo pulp regeneration. J Dent Res. 2011; 90:1013–1018.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Characterization of Human Dental Pulp Cells from Supernumerary Teeth by Using Flow Cytometry Analysis
- Methods of Isolation and Characterization of Stem Cells from Different Regions of Oral Cavity Using Markers: A Systematic Review
- Comparison of Gene Expression from Supernumerary Dental Pulp and Periodontal Ligament Stem Cells
- Dental Pulp Stem Cells and Current in vivo Approaches to Study Dental Pulp Stem Cells in Pulp Injury and Regeneration
- Dlx3 and Dlx5 Inhibit Adipogenic Differentiation of Human Dental Pulp Stem Cells