J Breast Cancer.
2016 Jun;19(2):148-155. 10.4048/jbc.2016.19.2.148.
Aberrant Expression of Breast Development-Related MicroRNAs, miR-22, miR-132, and miR-212, in Breast Tumor Tissues
- Affiliations
-
- 1Department of Genetics, Faculty of Biological Sciences, Tarbiat Modares University, Tehran, Iran. sjmowla@modares.ac.ir
- 2Department of Genetics, Faculty of Biological Sciences, Science and Research Branch, Islamic Azad University, Tehran, Iran.
- 3Razi Drug Research Center, Iran University of Medical Sciences, Tehran, Iran.
- 4Pathology Laboratory, Shariati Hospital, Tehran University of Medical Sciences, Tehran, Iran.
- 5Human Genetic Research Center, Baqiyatallah University of Medical Sciences, Tehran, Iran.
- KMID: 2308965
- DOI: http://doi.org/10.4048/jbc.2016.19.2.148
Abstract
- PURPOSE
MicroRNAs (miRNAs) are a major class of small endogenous RNA molecules that posttranscriptionally regulate the expression of most genes in the human genome. miRNAs are often located in chromosomal fragile sites, which are suscept-ible to amplification or deletion. Chromosomal deletions are frequent events in breast cancer cells. Deletion and loss of heterozygosity at 17p13.3 have been reported in 49% of breast cancers. The aim of the current study was to evaluate potential expression alterations of miR-22, miR-132, and miR-212, which are located on the 17p13.3 locus and are required for mammary gland development.
METHODS
A matched case-control study was conducted, which included 36 pairs of tumor and matched nontumor surgical specimens from patients diagnosed with breast invasive ductal carcinoma. Formalin-fixed paraffin-embedded samples from archival collections at the pathology department of Shariati Hospital were prepared for RNA extraction using the xylene-ethanol method before total RNA was isolated with TRIzol Reagent. Specific primers were designed for cDNA synthesis and miRNA amplification. The expression of miRNAs was then evaluated by real-time polymerase chain reaction (RT-PCR).
RESULTS
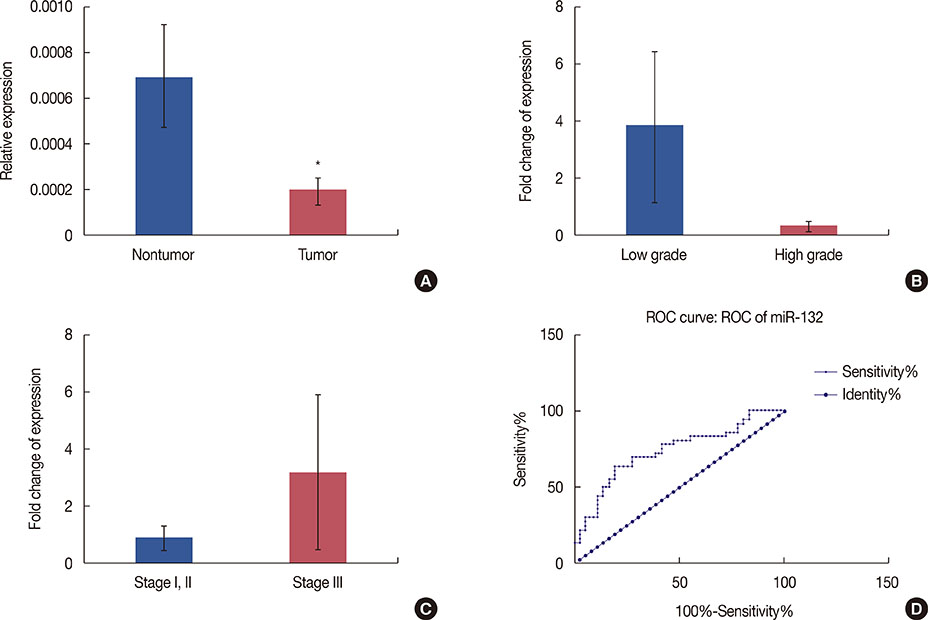
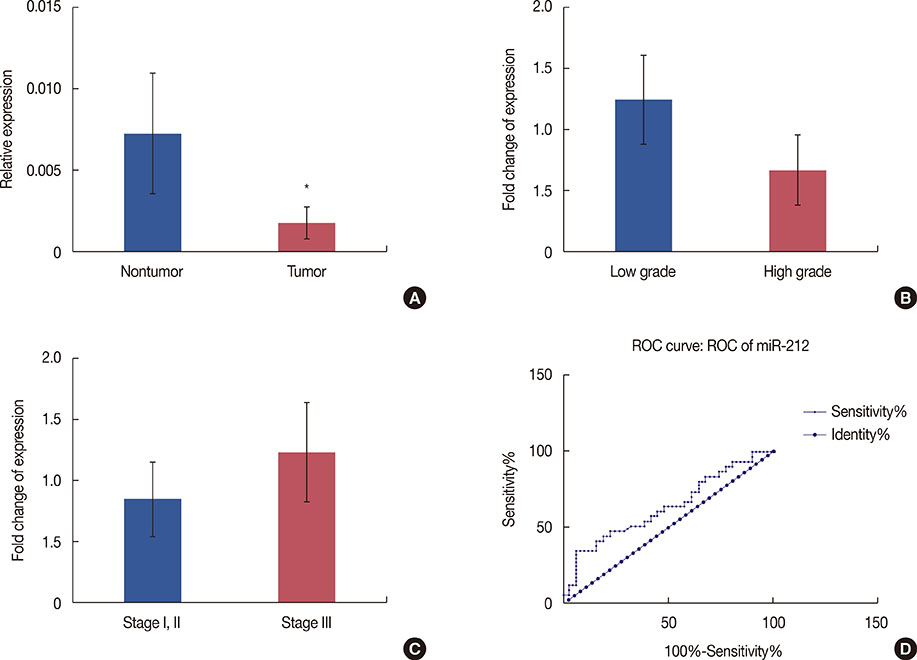
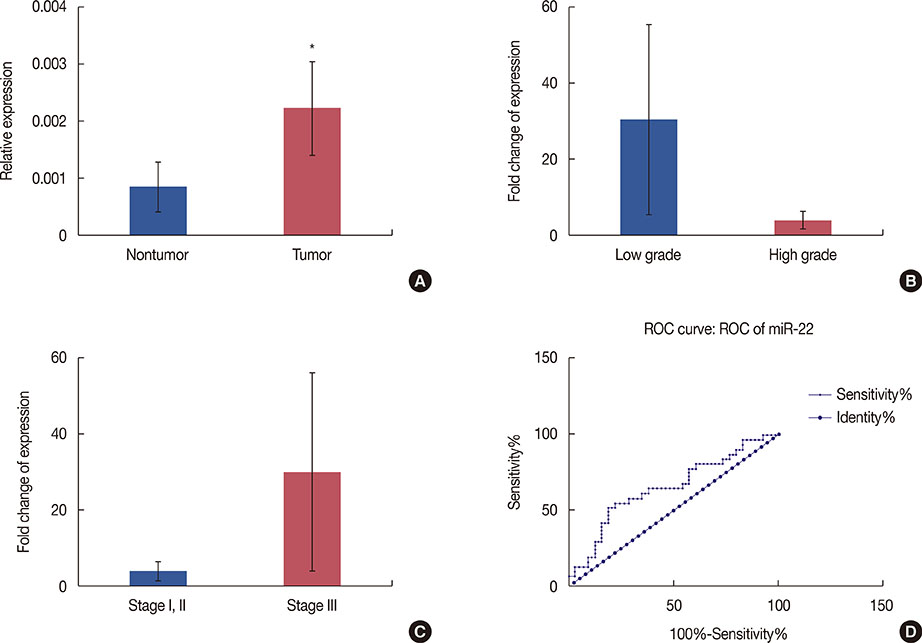
According to our RT-PCR data, the miR-212/miR-132 family was downregulated in breast cancer (0.328-fold, p<0.001), and this reduced expression was the most prominent in high-grade tumors. In contrast, miR-22 exhibited a significant upregulation in breast tumor samples (2.183-fold, p=0.040).
CONCLUSION
Consistent with the frequent deletion of the 17p13.3 locus in breast tumor cells, our gene expression data demonstrated a significant downregulation of miR-212 and miR-132 in breast cancer tissues. In contrast, we observed a significant upregulation of miR-22 in breast tumor samples. The latter conflicting result may have been due to the upregulation of miR-22 in stromal/cancer-associated fibroblasts, rather than in the tumor cells.
Keyword
MeSH Terms
-
Biomarkers
Breast Neoplasms*
Breast*
Carcinoma, Ductal
Case-Control Studies
Chromosome Deletion
DNA, Complementary
Down-Regulation
Fibroblasts
Gene Expression
Genome, Human
Humans
Loss of Heterozygosity
Mammary Glands, Human
Methods
MicroRNAs*
Pathology
Real-Time Polymerase Chain Reaction
RNA
Up-Regulation
Biomarkers
DNA, Complementary
MicroRNAs
RNA
Figure
Reference
-
1. Iorio MV, Casalini P, Piovan C, Braccioli L, Tagliabue E. Breast cancer and microRNAs: therapeutic impact. Breast. 2011; 20:Suppl 3. S63–S70.
Article2. Lerebours F, Lidereau R. Molecular alterations in sporadic breast cancer. Crit Rev Oncol Hematol. 2002; 44:121–141.
Article3. Kashiwagi H, Uchida K. Genome-wide profiling of gene amplification and deletion in cancer. Hum Cell. 2000; 13:135–141.4. Cornelis RS, van Vliet M, Vos CB, Cleton-Jansen AM, van de Vijver MJ, Peterse JL, et al. Evidence for a gene on 17p13.3, distal to TP53, as a target for allele loss in breast tumors without p53 mutations. Cancer Res. 1994; 54:4200–4206.
Article5. Isomura M, Tanigami A, Saito H, Harada Y, Katagiri T, Inazawa J, et al. Detailed analysis of loss of heterozygosity on chromosome band 17p13 in breast carcinoma on the basis of a high-resolution physical map with 29 markers. Genes Chromosomes Cancer. 1994; 9:173–179.
Article6. Piao HL, Ma L. Non-coding RNAs as regulators of mammary development and breast cancer. J Mammary Gland Biol Neoplasia. 2012; 17:33–42.
Article7. Ucar A, Vafaizadeh V, Jarry H, Fiedler J, Klemmt PA, Thum T, et al. miR-212 and miR-132 are required for epithelial stromal interactions necessary for mouse mammary gland development. Nat Genet. 2010; 42:1101–1108.
Article8. Ibarra I, Erlich Y, Muthuswamy SK, Sachidanandam R, Hannon GJ. A role for microRNAs in maintenance of mouse mammary epithelial progenitor cells. Genes Dev. 2007; 21:3238–3243.
Article9. Yu Z, Baserga R, Chen L, Wang C, Lisanti MP, Pestell RG. MicroRNA, cell cycle, and human breast cancer. Am J Pathol. 2010; 176:1058–1064.
Article10. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993; 75:843–854.
Article11. Lee YS, Dutta A. MicroRNAs: small but potent oncogenes or tumor suppressors. Curr Opin Investig Drugs. 2006; 7:560–564.12. Gregory RI, Shiekhattar R. MicroRNA biogenesis and cancer. Cancer Res. 2005; 65:3509–3512.13. Wu Q, Li H, Lu J, Ge Q, Lu Z. Aberrant microRNA expression in the development of breast carcinoma. Chin Sci Bull. 2010; 55:3517–3526.
Article14. Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005; 65:7065–7070.
Article15. Zhang J, Yang Y, Yang T, Liu Y, Li A, Fu S, et al. MicroRNA-22, downregulated in hepatocellular carcinoma and correlated with prognosis, suppresses cell proliferation and tumourigenicity. Br J Cancer. 2010; 103:1215–1220.
Article16. Anand S, Majeti BK, Acevedo LM, Murphy EA, Mukthavaram R, Scheppke L, et al. MicroRNA-132-mediated loss of p120RasGAP activates the endothelium to facilitate pathological angiogenesis. Nat Med. 2010; 16:909–914.
Article17. Bar N, Dikstein R. miR-22 forms a regulatory loop in PTEN/AKT pathway and modulates signaling kinetics. PLoS One. 2010; 5:e10859.
Article18. Pandey DP, Picard D. miR-22 inhibits estrogen signaling by directly targeting the estrogen receptor alpha mRNA. Mol Cell Biol. 2009; 29:3783–3790.
Article19. Park JK, Henry JC, Jiang J, Esau C, Gusev Y, Lerner MR, et al. miR-132 and miR-212 are increased in pancreatic cancer and target the retinoblastoma tumor suppressor. Biochem Biophys Res Commun. 2011; 406:518–523.
Article20. Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, et al. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008; 40:43–50.
Article21. He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004; 5:522–531.
Article22. Zhang S, Hao J, Xie F, Hu X, Liu C, Tong J, et al. Downregulation of miR-132 by promoter methylation contributes to pancreatic cancer development. Carcinogenesis. 2011; 32:1183–1189.
Article23. Song SJ, Ito K, Ala U, Kats L, Webster K, Sun SM, et al. The oncogenic microRNA miR-22 targets the TET2 tumor suppressor to promote hematopoietic stem cell self-renewal and transformation. Cell Stem Cell. 2013; 13:87–101.
Article24. Xu D, Takeshita F, Hino Y, Fukunaga S, Kudo Y, Tamaki A, et al. miR-22 represses cancer progression by inducing cellular senescence. J Cell Biol. 2011; 193:409–424.
Article25. Tsuchiya N, Izumiya M, Ogata-Kawata H, Okamoto K, Fujiwara Y, Nakai M, et al. Tumor suppressor miR-22 determines p53-dependent cellular fate through post-transcriptional regulation of p21. Cancer Res. 2011; 71:4628–4639.
Article26. Jazbutyte V, Fiedler J, Kneitz S, Galuppo P, Just A, Holzmann A, et al. MicroRNA-22 increases senescence and activates cardiac fibroblasts in the aging heart. Age (Dordr). 2013; 35:747–762.
Article27. Nouraee N, Van Roosbroeck K, Vasei M, Semnani S, Samaei NM, Naghshvar F, et al. Expression, tissue distribution and function of miR-21 in esophageal squamous cell carcinoma. PLoS One. 2013; 8:e73009.
Article28. Nouraee N, Mowla SJ, Calin GA. Tracking miRNAs' footprints in tumor–microenvironment interactions: insights and implications for targeted cancer therapy. Genes Chromosomes Cancer. 2015; 54:335–352.
Article29. Hanieh H. Aryl hydrocarbon receptor-microRNA-212/132 axis in human breast cancer suppresses metastasis by targeting SOX4. Mol Cancer. 2015; 14:172.
Article30. Zhang ZG, Chen WX, Wu YH, Liang HF, Zhang BX. MiR-132 prohibits proliferation, invasion, migration, and metastasis in breast cancer by targeting HN1. Biochem Biophys Res Commun. 2014; 454:109–114.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- miR-205 and miR-200c: Predictive Micro RNAs for Lymph Node Metastasis in Triple Negative Breast Cancer
- The Significance of MicroRNA Let-7b, miR-30c, and miR-200c Expression in Breast Cancers
- Prognostic Implications of MicroRNA-21 Overexpression in Invasive Ductal Carcinomas of the Breast
- LncRNA DLG1-AS1 Promotes Cancer Cell Proliferation in Triple Negative Breast Cancer by Downregulating miR-203
- Quantitative Measurement of Serum MicroRNA-21 Expression in Relation to Breast Cancer Metastasis in Chinese Females