J Breast Cancer.
2011 Dec;14(4):269-275. 10.4048/jbc.2011.14.4.269.
Prognostic Implications of MicroRNA-21 Overexpression in Invasive Ductal Carcinomas of the Breast
- Affiliations
-
- 1Department of Surgery, Korea University College of Medicine, Seoul, Korea. kujwbae@korea.ac.kr
- 2Center for Breast Cancer, Research Institute and Hospital, National Cancer Center, Goyang, Korea.
- 3Department of Pathology, Korea University College of Medicine, Seoul, Korea.
- KMID: 2175719
- DOI: http://doi.org/10.4048/jbc.2011.14.4.269
Abstract
- PURPOSE
Among more than 500 microRNAs, microRNA-21 (miR-21) is known to act as an oncogene. The aim of this study was to investigate the significance of miR-21 expression level in relation with clinicopathological factors and prognosis in breast cancer.
METHODS
MicroRNA was extracted from cancer and normal breast tissue of 109 breast cancer patients who underwent surgery from 2002 to 2004 using the Taqman(R) MicroRNA Assay. The correlation between miR-21 expression and clinicopathologic features was analyzed and the significance of miR-21 as a prognostic factor and its relationship with survival was determined.
RESULTS
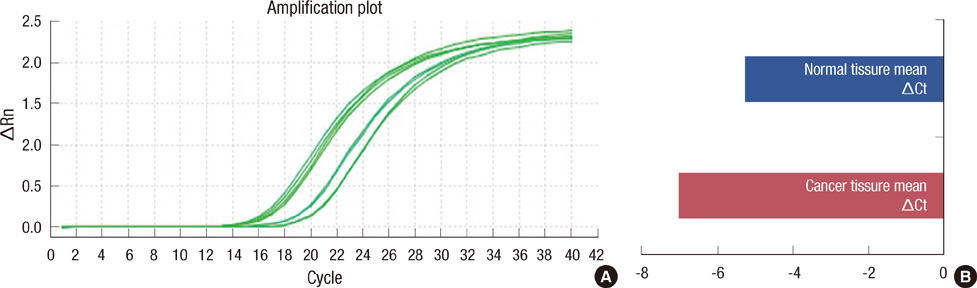
MiR-21 expression was higher in cancer tissues than in normal tissues (p<0.0001). High miR-21 expression was associated with mastectomy, larger tumor size, higher stage, higher grade, estrogen receptor (ER) negative, human epidermal growth factor receptor 2 (HER2) positive, HER2 positive breast cancer subtype, high Ki-67 expression, and death. On multivariate analysis, prognostic factors for overall survival were ER and miR-21. High miR-21 expression was significantly related to lower overall survival (p=0.031).
CONCLUSION
This study supports the role of miR-21 as an oncogene and a biomarker for breast cancer with its high expression in cancer tissues and its relationship with other prognostic factors and survival.
Keyword
MeSH Terms
Figure
Cited by 1 articles
-
miR-205 and miR-200c: Predictive Micro RNAs for Lymph Node Metastasis in Triple Negative Breast Cancer
Ufuk Berber, Ismail Yilmaz, Gizem Narli, Aptullah Haholu, Zafer Kucukodaci, Dilaver Demirel
J Breast Cancer. 2014;17(2):143-148. doi: 10.4048/jbc.2014.17.2.143.
Reference
-
1. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993. 75:843–854.
Article2. Lowery AJ, Miller N, McNeill RE, Kerin MJ. MicroRNAs as prognostic indicators and therapeutic targets: potential effect on breast cancer management. Clin Cancer Res. 2008. 14:360–365.
Article3. Verghese ET, Hanby AM, Speirs V, Hughes TA. Small is beautiful: microRNAs and breast cancer-where are we now? J Pathol. 2008. 215:214–221.
Article4. Shi M, Guo N. MicroRNA expression and its implications for the diagnosis and therapeutic strategies of breast cancer. Cancer Treat Rev. 2009. 35:328–334.
Article5. Lynam-Lennon N, Maher SG, Reynolds JV. The roles of microRNA in cancer and apoptosis. Biol Rev Camb Philos Soc. 2009. 84:55–71.
Article6. Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006. 6:857–866.
Article7. Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005. 65:7065–7070.
Article8. Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007. 26:2799–2803.
Article9. Qian B, Katsaros D, Lu L, Preti M, Durando A, Arisio R, et al. High miR-21 expression in breast cancer associated with poor disease-free survival in early stage disease and high TGF-beta1. Breast Cancer Res Treat. 2009. 117:131–140.
Article10. Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, et al. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008. 14:2348–2360.
Article11. Huang GL, Zhang XH, Guo GL, Huang KT, Yang KY, Shen X, et al. Clinical significance of miR-21 expression in breast cancer: SYBR-Green I-based real-time RT-PCR study of invasive ductal carcinoma. Oncol Rep. 2009. 21:673–679.
Article12. Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem. 2008. 283:1026–1033.
Article13. Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1). J Biol Chem. 2007. 282:14328–14336.
Article14. Edge SB, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A. AJCC Cancer Staging Manual. 2010. 7th ed. New York: Springer;347–376.15. Yu Z, Baserga R, Chen L, Wang C, Lisanti MP, Pestell RG. microRNA, cell cycle, and human breast cancer. Am J Pathol. 2010. 176:1058–1064.
Article16. Heneghan HM, Miller N, Lowery AJ, Sweeney KJ, Kerin MJ. MicroRNAs as novel biomarkers for breast cancer. J Oncol. 2009. 2009:950201.
Article17. Zoon CK, Starker EQ, Wilson AM, Emmert-Buck MR, Libutti SK, Tangrea MA. Current molecular diagnostics of breast cancer and the potential incorporation of microRNA. Expert Rev Mol Diagn. 2009. 9:455–467.
Article18. Lankat-Buttgereit B, Göke R. Programmed cell death protein 4 (pdcd4): a novel target for antineoplastic therapy? Biol Cell. 2003. 95:515–519.
Article19. Buzdar A. The place of chemotherapy in the treatment of early breast cancer. Br J Cancer. 1998. 78:Suppl 4. 16–20.
Article20. Dowsett M, Allred C, Knox J, Quinn E, Salter J, Wale C, et al. Relationship between quantitative estrogen and progesterone receptor expression and human epidermal growth factor receptor 2 (HER-2) status with recurrence in the Arimidex, Tamoxifen, Alone or in Combination trial. J Clin Oncol. 2008. 26:1059–1065.
Article21. Paik S, Hazan R, Fisher ER, Sass RE, Fisher B, Redmond C, et al. Pathologic findings from the National Surgical Adjuvant Breast and Bowel Project: prognostic significance of erbB-2 protein overexpression in primary breast cancer. J Clin Oncol. 1990. 8:103–112.
Article22. Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006. 295:2492–2502.
Article23. van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002. 415:530–536.24. Lowery AJ, Miller N, Devaney A, McNeill RE, Davoren PA, Lemetre C, et al. MicroRNA signatures predict oestrogen receptor, progesterone receptor and HER2/neu receptor status in breast cancer. Breast Cancer Res. 2009. 11:R27.25. Farragher SM, Tanney A, Kennedy RD, Paul Harkin D. RNA expression analysis from formalin fixed paraffin embedded tissues. Histochem Cell Biol. 2008. 130:435–445.
Article26. Doleshal M, Magotra AA, Choudhury B, Cannon BD, Labourier E, Szafranska AE. Evaluation and validation of total RNA extraction methods for microRNA expression analyses in formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2008. 10:203–211.
Article27. Li J, Smyth P, Flavin R, Cahill S, Denning K, Aherne S, et al. Comparison of miRNA expression patterns using total RNA extracted from matched samples of formalin-fixed paraffin-embedded (FFPE) cells and snap frozen cells. BMC Biotechnol. 2007. 7:36.
Article28. Siebolts U, Varnholt H, Drebber U, Dienes HP, Wickenhauser C, Odenthal M. Tissues from routine pathology archives are suitable for microRNA analyses by quantitative PCR. J Clin Pathol. 2009. 62:84–88.
Article29. Hasemeier B, Christgen M, Kreipe H, Lehmann U. Reliable microRNA profiling in routinely processed formalin-fixed paraffin-embedded breast cancer specimens using fluorescence labelled bead technology. BMC Biotechnol. 2008. 8:90.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Expression of Survivin According to Malignant Progression of Breast Lesions
- Cyclin D1 Expression in 101 Cases of Breast Carcinoma
- Expression of COX-2 in Cells of Invasive Ductal Carcinoma and Adjacent Non-Cancerous Ductal Epithelia in Human Breast
- Comparing Fluorescence In Situ Hybridization and Immunohistochemistry to Determine the HER-2/neu Status in Breast Carcinoma
- Invasive Ductal Carcinoma of the Male Breast: A Case Report and Review of the Literature