Ann Dermatol.
2013 Feb;25(1):36-45. 10.5021/ad.2013.25.1.36.
Comparison of Gene Expression Profiles between Keratinocytes, Melanocytes and Fibroblasts
- Affiliations
-
- 1Department of Dermatology, Research Institute for Medical Sciences, School of Medicine, Chungnam National University, Daejeon, Korea.
- 2Department of Dermatology, School of Medicine, Medical Research Institute, Chungbuk National University, Cheongju, Korea. jyl@chungbuk.ac.kr
- KMID: 2265969
- DOI: http://doi.org/10.5021/ad.2013.25.1.36
Abstract
- BACKGROUND
The skin has many important functions such as protection, preservation, temperature regulation, and vitamin D synthesis. It is composed of a variety of cell types including keratinocytes, melanocytes and fibroblasts.
OBJECTIVE
We attempted to compare the gene expression profiles between keratinocytes, melanocytes and fibroblast, using cDNA microarray.
METHODS
Keratinocytes, melanocytes and fibroblasts were primary cultured from five foreskin specimens. Total RNAs were extracted and pooled to reduce the individual variations, and then used for cDNA microarray.
RESULTS
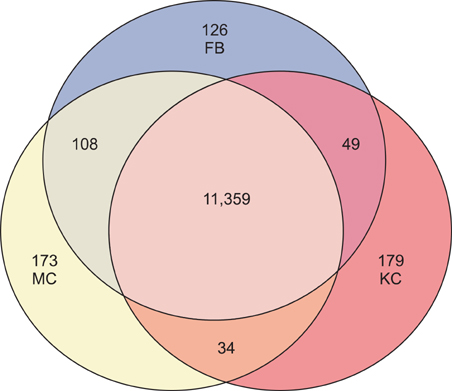
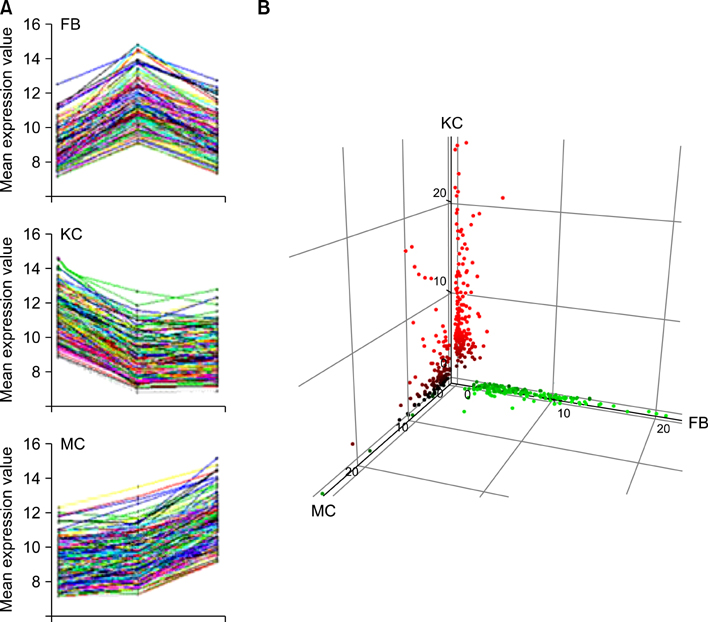
Total 12,028 genes were selected as the reliable genes whose expression was detected in at least one of the three cell types. By comparing the relative expression levels with cutoff limitation as a fourfold change, we obtained 126 fibroblast-specific, 179 keratinocyte-specific and 173 melanocyte-specific genes, many of which are known to be characteristically expressed in each cell type. In addition, we identified many genes whose skin-specific functions have not yet been determined.
CONCLUSION
Our data provide important information on which to base further investigation into the specification of skin cell types.
Keyword
MeSH Terms
Figure
Cited by 1 articles
-
Possible Role of Single Stranded DNA Binding Protein 3 on Skin Hydration by Regulating Epidermal Differentiation
Mi-Ra Choi, Jung-Min Shin, Young-Ah Shin, Yun-Hee Chang, Min-Youl Chang, Cho-Ah Lim, Kyung-Cheol Sohn, Young-Joon Seo, Chang-Deok Kim, Jeung-Hoon Lee, Young Lee
Ann Dermatol. 2018;30(4):432-440. doi: 10.5021/ad.2018.30.4.432.
Reference
-
1. Fartasch M, Ponec M. Improved barrier structure formation in air-exposed human keratinocyte culture systems. J Invest Dermatol. 1994. 102:366–374.
Article2. Sturm RA, Box NF, Ramsay M. Human pigmentation genetics: the difference is only skin deep. Bioessays. 1998. 20:712–721.
Article3. Takeda K, Gosiewska A, Peterkofsky B. Similar, but not identical, modulation of expression of extracellular matrix components during in vitro and in vivo aging of human skin fibroblasts. J Cell Physiol. 1992. 153:450–459.
Article4. Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995. 270:467–470.
Article5. Chee M, Yang R, Hubbell E, Berno A, Huang XC, Stern D, et al. Accessing genetic information with high-density DNA arrays. Science. 1996. 274:610–614.
Article6. Duggan DJ, Bittner M, Chen Y, Meltzer P, Trent JM. Expression profiling using cDNA microarrays. Nat Genet. 1999. 21:1 Suppl. 10–14.
Article7. Cheung VG, Morley M, Aguilar F, Massimi A, Kucherlapati R, Childs G. Making and reading microarrays. Nat Genet. 1999. 21:1 Suppl. 15–19.
Article8. Hwang C, Jang S, Choi DK, Kim S, Lee JH, Lee Y, et al. The role of nkx2.5 in keratinocyte differentiation. Ann Dermatol. 2009. 21:376–381.
Article9. Kim JH, Sohn KC, Choi TY, Kim MY, Ando H, Choi SJ, et al. Beta-catenin regulates melanocyte dendricity through the modulation of PKCzeta and PKCdelta. Pigment Cell Melanoma Res. 2010. 23:385–393.
Article10. Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, et al. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002. 30:e15.
Article11. Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998. 95:14863–14868.
Article12. Kerr MK, Martin M, Churchill GA. Analysis of variance for gene expression microarray data. J Comput Biol. 2000. 7:819–837.
Article13. Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990. 9:811–818.
Article14. Beissbarth T, Speed TP. GOstat: find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics. 2004. 20:1464–1465.
Article15. Pieraggi MT, Bouissou H, Angelier C, Uhart D, Magnol JP, Kokolo J. The fibroblast. Ann Pathol. 1985. 5:65–76.16. Hadshiew IM, Eller MS, Gilchrest BA. Skin aging and photoaging: the role of DNA damage and repair. Am J Contact Dermat. 2000. 11:19–25.
Article17. Yaar M, Gilchrest BA. Cellular and molecular mechanisms of cutaneous aging. J Dermatol Surg Oncol. 1990. 16:915–922.
Article18. Mundlos S, Chan D, Weng YM, Sillence DO, Cole WG, Bateman JF. Multiexon deletions in the type I collagen COL1A2 gene in osteogenesis imperfecta type IB. Molecules containing the shortened alpha2(I) chains show differential incorporation into the bone and skin extracellular matrix. J Biol Chem. 1996. 271:21068–21074.
Article19. Byers PH. Inherited disorders of collagen gene structure and expression. Am J Med Genet. 1989. 34:72–80.
Article20. Keene DR, Engvall E, Glanville RW. Ultrastructure of type VI collagen in human skin and cartilage suggests an anchoring function for this filamentous network. J Cell Biol. 1988. 107:1995–2006.
Article21. Scacheri PC, Gillanders EM, Subramony SH, Vedanarayanan V, Crowe CA, Thakore N, et al. Novel mutations in collagen VI genes: expansion of the Bethlem myopathy phenotype. Neurology. 2002. 58:593–602.
Article22. Epstein EH Jr. (Alpha1(3))3 human skin collagen. Release by pepsin digestion and preponderance in fetal life. J Biol Chem. 1974. 249:3225–3231.23. Prockop DJ, Sieron AL, Li SW. Procollagen N-proteinase and procollagen C-proteinase. Two unusual metalloproteinases that are essential for procollagen processing probably have important roles in development and cell signaling. Matrix Biol. 1998. 16:399–408.
Article24. Funderburgh JL, Funderburgh ML, Brown SJ, Vergnes JP, Hassell JR, Mann MM, et al. Sequence and structural implications of a bovine corneal keratan sulfate proteoglycan core protein. Protein 37B represents bovine lumican and proteins 37A and 25 are unique. J Biol Chem. 1993. 268:11874–11880.
Article25. Jiang CK, Tomić-Canić M, Lucas DJ, Simon M, Blumenberg M. TGF beta promotes the basal phenotype of epidermal keratinocytes: transcriptional induction of K#5 and K#14 keratin genes. Growth Factors. 1995. 12:87–97.
Article26. Lane EB, Rugg EL, Navsaria H, Leigh IM, Heagerty AH, Ishida-Yamamoto A, et al. A mutation in the conserved helix termination peptide of keratin 5 in hereditary skin blistering. Nature. 1992. 356:244–246.
Article27. Chen H, Bonifas JM, Matsumura K, Ikeda S, Leyden WA, Epstein EH Jr. Keratin 14 gene mutations in patients with epidermolysis bullosa simplex. J Invest Dermatol. 1995. 105:629–632.
Article28. Mischke D, Korge BP, Marenholz I, Volz A, Ziegler A. Genes encoding structural proteins of epidermal cornification and S100 calcium-binding proteins form a gene complex ("epidermal differentiation complex") on human chromosome 1q21. J Invest Dermatol. 1996. 106:989–992.
Article29. Gerke V, Moss SE. Annexins: from structure to function. Physiol Rev. 2002. 82:331–371.
Article30. Tait JF, Sakata M, McMullen BA, Miao CH, Funakoshi T, Hendrickson LE, et al. Placental anticoagulant proteins: isolation and comparative characterization four members of the lipocortin family. Biochemistry. 1988. 27:6268–6276.
Article31. Ernst JD, Hoye E, Blackwood RA, Jaye D. Purification and characterization of an abundant cytosolic protein from human neutrophils that promotes Ca2(+)-dependent aggregation of isolated specific granules. J Clin Invest. 1990. 85:1065–1071.
Article32. Thorey IS, Roth J, Regenbogen J, Halle JP, Bittner M, Vogl T, et al. The Ca2+-binding proteins S100A8 and S100A9 are encoded by novel injury-regulated genes. J Biol Chem. 2001. 276:35818–35825.
Article33. Ma AS, Ozers LJ. Annexins I and II show differences in subcellular localization and differentiation-related changes in human epidermal keratinocytes. Arch Dermatol Res. 1996. 288:596–603.
Article34. Allen E, Yu QC, Fuchs E. Mice expressing a mutant desmosomal cadherin exhibit abnormalities in desmosomes, proliferation, and epidermal differentiation. J Cell Biol. 1996. 133:1367–1382.
Article35. Haftek M, Serre G, Mils V, Thivolet J. Immunocytochemical evidence for a possible role of cross-linked keratinocyte envelopes in stratum corneum cohesion. J Histochem Cytochem. 1991. 39:1531–1538.
Article36. Ruzzi L, Gagnoux-Palacios L, Pinola M, Belli S, Meneguzzi G, D'Alessio M, et al. A homozygous mutation in the integrin alpha6 gene in junctional epidermolysis bullosa with pyloric atresia. J Clin Invest. 1997. 99:2826–2831.
Article37. Vidal F, Aberdam D, Miquel C, Christiano AM, Pulkkinen L, Uitto J, et al. Integrin beta 4 mutations associated with junctional epidermolysis bullosa with pyloric atresia. Nat Genet. 1995. 10:229–234.
Article38. Ryan MC, Lee K, Miyashita Y, Carter WG. Targeted disruption of the LAMA3 gene in mice reveals abnormalities in survival and late stage differentiation of epithelial cells. J Cell Biol. 1999. 145:1309–1323.
Article39. Jackson IJ. Homologous pigmentation mutations in human, mouse and other model organisms. Hum Mol Genet. 1997. 6:1613–1624.
Article40. Hearing VJ, Jiménez M. Analysis of mammalian pigmentation at the molecular level. Pigment Cell Res. 1989. 2:75–85.
Article41. Fischer WH, Spiess J. Identification of a mammalian glutaminyl cyclase converting glutaminyl into pyroglutamyl peptides. Proc Natl Acad Sci U S A. 1987. 84:3628–3632.
Article42. Yanagawa S, van Leeuwen F, Wodarz A, Klingensmith J, Nusse R. The dishevelled protein is modified by wingless signaling in Drosophila. Genes Dev. 1995. 9:1087–1097.
Article43. Goding CR. Mitf from neural crest to melanoma: signal transduction and transcription in the melanocyte lineage. Genes Dev. 2000. 14:1712–1728.
Article44. Price ER, Fisher DE. Sensorineural deafness and pigmentation genes: melanocytes and the Mitf transcriptional network. Neuron. 2001. 30:15–18.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Expression of Heat Shock Protein 70 in Human Skin Cells as a Photoprotective Function after UV Exposure
- Effect of suction blister fluid and IFN-gamma on HLA-A,B,C and HLA-DR espression in cultures human melanocytes
- Effect of Radiation on Cultured Human Normal Keratinocytes and Melanocytes
- Establishment and Characterization of an In Vitro Model for Cholesteatoma
- The Effect of Supernatant from UVB - Irradiated Cultured Keratinocytes on the Growth , Melanin Content , and Tyrosinase Activity of Human Melanocyte