Eosinophil Development, Regulation of Eosinophil-Specific Genes, and Role of Eosinophils in the Pathogenesis of Asthma
- Affiliations
-
- 1Division of Molecular and Life Sciences, College of Science and Technology, Hanyang University, Ansan, Korea. iychu@hanyang.ac.kr
- KMID: 2260359
- DOI: http://doi.org/10.4168/aair.2012.4.2.68
Abstract
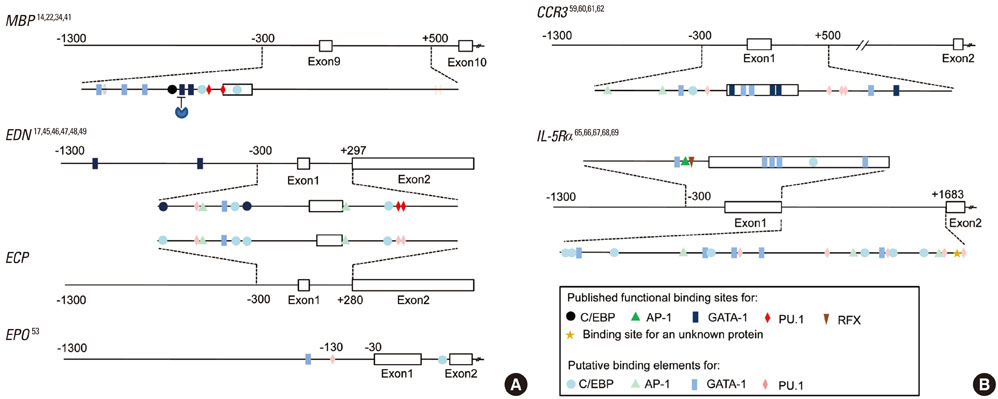
- Eosinophils arise from hematopoietic CD34+ stem cells in the bone marrow. They acquire IL-5Ralpha on their surface at a very early stage during eosinophilopoiesis, and differentiate under the strong influence of interleukin (IL)-5. They then exit to the bloodstream, and enter the lung upon exposure to airway inflammatory signals, including eotaxins. In inflamed tissues, eosinophils act as key mediators of terminal effector functions and innate immunity and in linking to adaptive immune responses. Transcription factors GATA-1, CCAAT/enhancer-binding protein, and PU.1 play instructive roles in eosinophil specification from multipotent stem cells through a network of cooperative and antagonistic interactions. Not surprisingly, the interplay of these transcription factors is instrumental in forming the regulatory circuit of expression of eosinophil-specific genes, encoding eosinophil major basic protein and neurotoxin, CC chemokine receptor 3 eotaxin receptor, and IL-5 receptor alpha. Interestingly, a common feature is that the critical cis-acting elements for these transcription factors are clustered in exon 1 and intron 1 of these genes rather than their promoters. Elucidation of the mechanism of eosinophil development and activation may lead to selective elimination of eosinophils in animals and human subjects. Furthermore, availability of a range of genetically modified mice lacking or overproducing eosinophil-specific genes will facilitate evaluation of the roles of eosinophils in the pathogenesis of asthma. This review summarizes eosinophil biology, focusing on development and regulation of eosinophil-specific genes, with a heavy emphasis on the causative link between eosinophils and pathological development of asthma using genetically modified mice as models of asthma.
MeSH Terms
-
Aluminum Hydroxide
Animals
Asthma
Biology
Bone Marrow
Carbonates
Eosinophil Major Basic Protein
Eosinophils
Exons
Humans
Immunity, Innate
Interleukin-5
Interleukins
Introns
Lung
Mice
Multipotent Stem Cells
Receptors, CCR3
Stem Cells
Transcription Factors
Aluminum Hydroxide
Carbonates
Eosinophil Major Basic Protein
Interleukin-5
Interleukins
Receptors, CCR3
Transcription Factors
Figure
Cited by 3 articles
-
Terminally Differentiating Eosinophils Express Neutrophil Primary Granule Proteins as well as Eosinophil-specific Granule Proteins in a Temporal Manner
Karam Kim, Sae Mi Hwang, Sung Min Kim, Sung Woo Park, Yunjae Jung, Il Yup Chung
Immune Netw. 2017;17(6):410-423. doi: 10.4110/in.2017.17.6.410.Hypereosinophilia-associated Diseases and the Therapeutic Agents in Development
Il Yup Chung
Hanyang Med Rev. 2013;33(1):65-74. doi: 10.7599/hmr.2013.33.1.65.The prevalence of bronchial hyperresponsiveness in elementary school children and its associated factors
Mi Suk Kim, Young Ho Kim, Dong In Suh, Young Yull Koh, Byoung-Ju Kim, Hyo Bin Kim, So Yeon Lee, Dae Jin Song, Woo-Kyung Kim, Gwang Cheon Jang, Jung Yeon Shim, Soo-Jong Hong, Ji-Won Kwon
Allergy Asthma Respir Dis. 2014;2(3):171-178. doi: 10.4168/aard.2014.2.3.171.
Reference
-
1. Young B, Lowe JS, Stevens A, Heath JW, Heath JW. Wheater's functional histology: a text and colour atlas. 2006. 5th ed. Edinburgh: Elsevier.2. Weller PF, Dvorak AM. Busse W, Holgate ST, editors. Human eosinophils-development, maturation and functional morphology. Asthma and rhinitis. 1994. Boston: Blackwell Scientific;225–274.3. Ramji DP, Foka P. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem J. 2002. 365:561–575.4. Müller C, Kowenz-Leutz E, Grieser-Ade S, Graf T, Leutz A. NF-M (chicken C/EBP beta) induces eosinophilic differentiation and apoptosis in a hematopoietic progenitor cell line. EMBO J. 1995. 14:6127–6135.5. Zhang DE, Zhang P, Wang ND, Hetherington CJ, Darlington GJ, Tenen DG. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein alpha-deficient mice. Proc Natl Acad Sci U S A. 1997. 94:569–574.6. Nerlov C, McNagny KM, Döderlein G, Kowenz-Leutz E, Graf T. Distinct C/EBP functions are required for eosinophil lineage commitment and maturation. Genes Dev. 1998. 12:2413–2423.7. Iwama A, Osawa M, Hirasawa R, Uchiyama N, Kaneko S, Onodera M, Shibuya K, Shibuya A, Vinson C, Tenen DG, Nakauchi H. Reciprocal roles for CCAAT/enhancer binding protein (C/EBP) and PU.1 transcription factors in Langerhans cell commitment. J Exp Med. 2002. 195:547–558.8. Tanaka T, Akira S, Yoshida K, Umemoto M, Yoneda Y, Shirafuji N, Fujiwara H, Suematsu S, Yoshida N, Kishimoto T. Targeted disruption of the NF-IL6 gene discloses its essential role in bacteria killing and tumor cytotoxicity by macrophages. Cell. 1995. 80:353–361.9. Ferreira R, Ohneda K, Yamamoto M, Philipsen S. GATA1 function, a paradigm for transcription factors in hematopoiesis. Mol Cell Biol. 2005. 25:1215–1227.10. Kulessa H, Frampton J, Graf T. GATA-1 reprograms avian myelomonocytic cell lines into eosinophils, thromboblasts, and erythroblasts. Genes Dev. 1995. 9:1250–1262.11. Hirasawa R, Shimizu R, Takahashi S, Osawa M, Takayanagi S, Kato Y, Onodera M, Minegishi N, Yamamoto M, Fukao K, Taniguchi H, Nakauchi H, Iwama A. Essential and instructive roles of GATA factors in eosinophil development. J Exp Med. 2002. 195:1379–1386.12. Yu C, Cantor AB, Yang H, Browne C, Wells RA, Fujiwara Y, Orkin SH. Targeted deletion of a high-affinity GATA-binding site in the GATA-1 promoter leads to selective loss of the eosinophil lineage in vivo. J Exp Med. 2002. 195:1387–1395.13. Iwasaki H, Mizuno S, Mayfield R, Shigematsu H, Arinobu Y, Seed B, Gurish MF, Takatsu K, Akashi K. Identification of eosinophil lineage-committed progenitors in the murine bone marrow. J Exp Med. 2005. 201:1891–1897.14. Yamaguchi Y, Nishio H, Kishi K, Ackerman SJ, Suda T. C/EBPbeta and GATA-1 synergistically regulate activity of the eosinophil granule major basic protein promoter: implication for C/EBPbeta activity in eosinophil gene expression. Blood. 1999. 94:1429–1439.15. McNagny KM, Sieweke MH, Döderlein G, Graf T, Nerlov C. Regulation of eosinophil-specific gene expression by a C/EBP-Ets complex and GATA-1. EMBO J. 1998. 17:3669–3680.16. Iwasaki H, Mizuno S, Arinobu Y, Ozawa H, Mori Y, Shigematsu H, Takatsu K, Tenen DG, Akashi K. The order of expression of transcription factors directs hierarchical specification of hematopoietic lineages. Genes Dev. 2006. 20:3010–3021.17. Qiu Z, Dyer KD, Xie Z, Rådinger M, Rosenberg HF. GATA transcription factors regulate the expression of the human eosinophil-derived neurotoxin (RNase 2) gene. J Biol Chem. 2009. 284:13099–13109.18. Tsai FY, Orkin SH. Transcription factor GATA-2 is required for proliferation/survival of early hematopoietic cells and mast cell formation, but not for erythroid and myeloid terminal differentiation. Blood. 1997. 89:3636–3643.19. McNagny K, Graf T. Making eosinophils through subtle shifts in transcription factor expression. J Exp Med. 2002. 195:F43–F47.20. Klemsz MJ, McKercher SR, Celada A, Van Beveren C, Maki RA. The macrophage and B cell-specific transcription factor PU.1 is related to the ets oncogene. Cell. 1990. 61:113–124.21. Nerlov C, Graf T. PU.1 induces myeloid lineage commitment in multipotent hematopoietic progenitors. Genes Dev. 1998. 12:2403–2412.22. Gombart AF, Kwok SH, Anderson KL, Yamaguchi Y, Torbett BE, Koeffler HP. Regulation of neutrophil and eosinophil secondary granule gene expression by transcription factors C/EBP epsilon and PU.1. Blood. 2003. 101:3265–3273.23. Tsang AP, Visvader JE, Turner CA, Fujiwara Y, Yu C, Weiss MJ, Crossley M, Orkin SH. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell. 1997. 90:109–119.24. Querfurth E, Schuster M, Kulessa H, Crispino JD, Döderlein G, Orkin SH, Graf T, Nerlov C. Antagonism between C/EBPbeta and FOG in eosinophil lineage commitment of multipotent hematopoietic progenitors. Genes Dev. 2000. 14:2515–2525.25. Weisz A, Marx P, Sharf R, Appella E, Driggers PH, Ozato K, Levi BZ. Human interferon consensus sequence binding protein is a negative regulator of enhancer elements common to interferon-inducible genes. J Biol Chem. 1992. 267:25589–25596.26. Milanovic M, Terszowski G, Struck D, Liesenfeld O, Carstanjen D. IFN consensus sequence binding protein (Icsbp) is critical for eosinophil development. J Immunol. 2008. 181:5045–5053.27. Biggs J, Murphy EV, Israel MA. A human Id-like helix-loop-helix protein expressed during early development. Proc Natl Acad Sci U S A. 1992. 89:1512–1516.28. Buitenhuis M, van Deutekom HW, Verhagen LP, Castor A, Jacobsen SE, Lammers JW, Koenderman L, Coffer PJ. Differential regulation of granulopoiesis by the basic helix-loop-helix transcriptional inhibitors Id1 and Id2. Blood. 2005. 105:4272–4281.29. Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999. 284:770–776.30. Kang JH, Lee DH, Lee JS, Kim HJ, Shin JW, Lee YH, Lee YS, Park CS, Chung IY. Eosinophilic differentiation is promoted by blockage of Notch signaling with a gamma-secretase inhibitor. Eur J Immunol. 2005. 35:2982–2990.31. Kang JH, Lee da H, Seo H, Park JS, Nam KH, Shin SY, Park CS, Chung IY. Regulation of functional phenotypes of cord blood derived eosinophils by gamma-secretase inhibitor. Am J Respir Cell Mol Biol. 2007. 37:571–577.32. Radke AL, Reynolds LE, Melo RC, Dvorak AM, Weller PF, Spencer LA. Mature human eosinophils express functional Notch ligands mediating eosinophil autocrine regulation. Blood. 2009. 113:3092–3101.33. Diggs LW, Sturm D, Bell A. The morphology of human blood cells. 1975. Chicago: Abbott Laboratories.34. Du J, Stankiewicz MJ, Liu Y, Xi Q, Schmitz JE, Lekstrom-Himes JA, Ackerman SJ. Novel combinatorial interactions of GATA-1, PU.1, and C/EBPepsilon isoforms regulate transcription of the gene encoding eosinophil granule major basic protein. J Biol Chem. 2002. 277:43481–43494.35. Bedi R, Du J, Sharma AK, Gomes I, Ackerman SJ. Human C/EBP-epsilon activator and repressor isoforms differentially reprogram myeloid lineage commitment and differentiation. Blood. 2009. 113:317–327.36. Arinobu Y, Iwasaki H, Gurish MF, Mizuno S, Shigematsu H, Ozawa H, Tenen DG, Austen KF, Akashi K. Developmental checkpoints of the basophil/mast cell lineages in adult murine hematopoiesis. Proc Natl Acad Sci U S A. 2005. 102:18105–18110.37. Mori Y, Iwasaki H, Kohno K, Yoshimoto G, Kikushige Y, Okeda A, Uike N, Niiro H, Takenaka K, Nagafuji K, Miyamoto T, Harada M, Takatsu K, Akashi K. Identification of the human eosinophil lineage-committed progenitor: revision of phenotypic definition of the human common myeloid progenitor. J Exp Med. 2009. 206:183–193.38. Boyce JA, Friend D, Matsumoto R, Austen KF, Owen WF. Differentiation in vitro of hybrid eosinophil/basophil granulocytes: autocrine function of an eosinophil developmental intermediate. J Exp Med. 1995. 182:49–57.39. Byström J, Wynn TA, Domachowske JB, Rosenberg HF. Gene microarray analysis reveals interleukin-5-dependent transcriptional targets in mouse bone marrow. Blood. 2004. 103:868–877.40. Li MS, Sun L, Satoh T, Fisher LM, Spry CJ. Human eosinophil major basic protein, a mediator of allergic inflammation, is expressed by alternative splicing from two promoters. Biochem J. 1995. 305:921–927.41. Yamaguchi Y, Zon LI, Ackerman SJ, Yamamoto M, Suda T. Forced GATA-1 expression in the murine myeloid cell line M1: induction of c-Mpl expression and megakaryocytic/erythroid differentiation. Blood. 1998. 91:450–457.42. Plager DA, Adolphson CR, Gleich GJ. A novel human homolog of eosinophil major basic protein. Immunol Rev. 2001. 179:192–202.43. Gomolin HI, Yamaguchi Y, Paulpillai AV, Dvorak LA, Ackerman SJ, Tenen DG. Human eosinophil Charcot-Leyden crystal protein: cloning and characterization of a lysophospholipase gene promoter. Blood. 1993. 82:1868–1874.44. Yang D, Chen Q, Su SB, Zhang P, Kurosaka K, Caspi RR, Michalek SM, Rosenberg HF, Zhang N, Oppenheim JJ. Eosinophil-derived neurotoxin acts as an alarmin to activate the TLR2-MyD88 signal pathway in dendritic cells and enhances Th2 immune responses. J Exp Med. 2008. 205:79–90.45. Tiffany HL, Handen JS, Rosenberg HF. Enhanced expression of the eosinophil-derived neurotoxin ribonuclease (RNS2) gene requires interaction between the promoter and intron. J Biol Chem. 1996. 271:12387–12393.46. van Dijk TB, Caldenhoven E, Raaijmakers JA, Lammers JW, Koenderman L, de Groot RP. The role of transcription factor PU.1 in the activity of the intronic enhancer of the eosinophil-derived neurotoxin (RNS2) gene. Blood. 1998. 91:2126–2132.47. Baltus B, Buitenhuis M, van Dijk TB, Vinson C, Raaijmakers JA, Lammers JW, Koenderman L, de Groot RP. C/EBP regulates the promoter of the eosinophil-derived neurotoxin/RNS2 gene in human eosinophilic cells. J Leukoc Biol. 1999. 66:683–688.48. Dyer KD, Nitto T, Moreau JM, McDevitt AL, Rosenberg HF. Identification of a purine-rich intronic enhancer element in the mouse eosinophil-associated ribonuclease 2 (mEar 2) gene. Mamm Genome. 2004. 15:126–134.49. Wang HY, Ho PC, Lan CY, Chang MD. Transcriptional regulation of human eosinophil RNase2 by the liver-enriched hepatocyte nuclear factor 4. J Cell Biochem. 2009. 106:317–326.50. Slifman NR, Loegering DA, McKean DJ, Gleich GJ. Ribonuclease activity associated with human eosinophil-derived neurotoxin and eosinophil cationic protein. J Immunol. 1986. 137:2913–2917.51. Wang HY, Chang HT, Pai TW, Wu CI, Lee YH, Chang YH, Tai HL, Tang CY, Chou WY, Chang MD. Transcriptional regulation of human eosinophil RNases by an evolutionary-conserved sequence motif in primate genome. BMC Mol Biol. 2007. 8:89.52. Sakamaki K, Tomonaga M, Tsukui K, Nagata S. Molecular cloning and characterization of a chromosomal gene for human eosinophil peroxidase. J Biol Chem. 1989. 264:16828–16836.53. Yamaguchi Y, Zhang DE, Sun Z, Albee EA, Nagata S, Tenen DG, Ackerman SJ. Functional characterization of the promoter for the gene encoding human eosinophil peroxidase. J Biol Chem. 1994. 269:19410–19419.54. Daugherty BL, Siciliano SJ, DeMartino JA, Malkowitz L, Sirotina A, Springer MS. Cloning, expression, and characterization of the human eosinophil eotaxin receptor. J Exp Med. 1996. 183:2349–2354.55. Ponath PD, Qin S, Post TW, Wang J, Wu L, Gerard NP, Newman W, Gerard C, Mackay CR. Molecular cloning and characterization of a human eotaxin receptor expressed selectively on eosinophils. J Exp Med. 1996. 183:2437–2448.56. Sallusto F, Mackay CR, Lanzavecchia A. Selective expression of the eotaxin receptor CCR3 by human T helper 2 cells. Science. 1997. 277:2005–2007.57. Forsythe P, Befus AD. CCR3: a key to mast cell phenotypic and functional diversity? Am J Respir Cell Mol Biol. 2003. 28:405–409.58. Beck LA, Tancowny B, Brummet ME, Asaki SY, Curry SL, Penno MB, Foster M, Bahl A, Stellato C. Functional analysis of the chemokine receptor CCR3 on airway epithelial cells. J Immunol. 2006. 177:3344–3354.59. Zimmermann N, Daugherty BL, Kavanaugh JL, El-Awar FY, Moulton EA, Rothenberg ME. Analysis of the CC chemokine receptor 3 gene reveals a complex 5' exon organization, a functional role for untranslated exon 1, and a broadly active promoter with eosinophil-selective elements. Blood. 2000. 96:2346–2354.60. Scotet E, Schroeder S, Lanzavecchia A. Molecular regulation of CC-chemokine receptor 3 expression in human T helper 2 cells. Blood. 2001. 98:2568–2570.61. Vijh S, Dayhoff DE, Wang CE, Imam Z, Ehrenberg PK, Michael NL. Transcription regulation of human chemokine receptor CCR3: evidence for a rare TATA-less promoter structure conserved between drosophila and humans. Genomics. 2002. 80:86–95.62. Kim BS, Uhm TG, Lee SK, Lee SH, Kang JH, Park CS, Chung IY. The crucial role of GATA-1 in CCR3 gene transcription: modulated balance by multiple GATA elements in the CCR3 regulatory region. J Immunol. 2010. 185:6866–6875.63. Kouro T, Takatsu K. IL-5- and eosinophil-mediated inflammation: from discovery to therapy. Int Immunol. 2009. 21:1303–1309.64. Rothenberg ME, Hogan SP. The eosinophil. Annu Rev Immunol. 2006. 24:147–174.65. Sun Z, Yergeau DA, Tuypens T, Tavernier J, Paul CC, Baumann MA, Tenen DG, Ackerman SJ. Identification and characterization of a functional promoter region in the human eosinophil IL-5 receptor alpha subunit gene. J Biol Chem. 1995. 270:1462–1471.66. Iwama A, Pan J, Zhang P, Reith W, Mach B, Tenen DG, Sun Z. Dimeric RFX proteins contribute to the activity and lineage specificity of the interleukin-5 receptor alpha promoter through activation and repression domains. Mol Cell Biol. 1999. 19:3940–3950.67. Baltus B, van Dijk TB, Caldenhoven E, Zanders E, Raaijmakers JA, Lammers JW, Koenderman L, de Groot RP. An AP-1 site in the promoter of the human IL-5R alpha gene is necessary for promoter activity in eosinophilic HL60 cells. FEBS Lett. 1998. 434:251–254.68. Emslie D, D'Costa K, Hasbold J, Metcalf D, Takatsu K, Hodgkin PO, Corcoran LM. Oct2 enhances antibody-secreting cell differentiation through regulation of IL-5 receptor alpha chain expression on activated B cells. J Exp Med. 2008. 205:409–421.69. Zhang J, Kuvelkar R, Cheewatrakoolpong B, Williams S, Egan RW, Billah MM. Evidence for multiple promoters of the human IL-5 receptor alpha subunit gene: a novel 6-base pair element determines cell-specific promoter function. J Immunol. 1997. 159:5412–5421.70. Adcock IM, Tsaprouni L, Bhavsar P, Ito K. Epigenetic regulation of airway inflammation. Curr Opin Immunol. 2007. 19:694–700.71. Bousquet J, Chanez P, Lacoste JY, Barnéon G, Ghavanian N, Enander I, Venge P, Ahlstedt S, Simony-Lafontaine J, Godard P, Michel FB. Eosinophilic inflammation in asthma. N Engl J Med. 1990. 323:1033–1039.72. Humbles AA, Lloyd CM, McMillan SJ, Friend DS, Xanthou G, McKenna EE, Ghiran S, Gerard NP, Yu C, Orkin SH, Gerard C. A critical role for eosinophils in allergic airways remodeling. Science. 2004. 305:1776–1779.73. Walsh ER, Sahu N, Kearley J, Benjamin E, Kang BH, Humbles A, August A. Strain-specific requirement for eosinophils in the recruitment of T cells to the lung during the development of allergic asthma. J Exp Med. 2008. 205:1285–1292.74. Lee JJ, Dimina D, Macias MP, Ochkur SI, McGarry MP, O'Neill KR, Protheroe C, Pero R, Nguyen T, Cormier SA, Lenkiewicz E, Colbert D, Rinaldi L, Ackerman SJ, Irvin CG, Lee NA. Defining a link with asthma in mice congenitally deficient in eosinophils. Science. 2004. 305:1773–1776.75. Jacobsen EA, Ochkur SI, Pero RS, Taranova AG, Protheroe CA, Colbert DC, Lee NA, Lee JJ. Allergic pulmonary inflammation in mice is dependent on eosinophil-induced recruitment of effector T cells. J Exp Med. 2008. 205:699–710.76. Robinson DS, Hamid Q, Ying S, Tsicopoulos A, Barkans J, Bentley AM, Corrigan C, Durham SR, Kay AB. Predominant TH2-like bronchoalveolar T-lymphocyte population in atopic asthma. N Engl J Med. 1992. 326:298–304.77. Sur S, Gleich GJ, Swanson MC, Bartemes KR, Broide DH. Eosinophilic inflammation is associated with elevation of interleukin-5 in the airways of patients with spontaneous symptomatic asthma. J Allergy Clin Immunol. 1995. 96:661–668.78. Foster PS, Hogan SP, Ramsay AJ, Matthaei KI, Young IG. Interleukin 5 deficiency abolishes eosinophilia, airways hyperreactivity, and lung damage in a mouse asthma model. J Exp Med. 1996. 183:195–201.79. Hamelmann E, Oshiba A, Loader J, Larsen GL, Gleich G, Lee J, Gelfand EW. Antiinterleukin-5 antibody prevents airway hyperresponsiveness in a murine model of airway sensitization. Am J Respir Crit Care Med. 1997. 155:819–825.80. Cho JY, Miller M, Baek KJ, Han JW, Nayar J, Lee SY, McElwain K, McElwain S, Friedman S, Broide DH. Inhibition of airway remodeling in IL-5-deficient mice. J Clin Invest. 2004. 113:551–560.81. Hogan SP, Matthaei KI, Young JM, Koskinen A, Young IG, Foster PS. A novel T cell-regulated mechanism modulating allergen-induced airways hyperreactivity in BALB/c mice independently of IL-4 and IL-5. J Immunol. 1998. 161:1501–1509.82. Mattes J, Yang M, Mahalingam S, Kuehr J, Webb DC, Simson L, Hogan SP, Koskinen A, McKenzie AN, Dent LA, Rothenberg ME, Matthaei KI, Young IG, Foster PS. Intrinsic defect in T cell production of interleukin (IL)-13 in the absence of both IL-5 and eotaxin precludes the development of eosinophilia and airways hyperreactivity in experimental asthma. J Exp Med. 2002. 195:1433–1444.83. Lee JJ, McGarry MP, Farmer SC, Denzler KL, Larson KA, Carrigan PE, Brenneise IE, Horton MA, Haczku A, Gelfand EW, Leikauf GD, Lee NA. Interleukin-5 expression in the lung epithelium of transgenic mice leads to pulmonary changes pathognomonic of asthma. J Exp Med. 1997. 185:2143–2156.84. Tanaka H, Kawada N, Yamada T, Kawada K, Takatsu K, Nagai H. Allergen-induced airway inflammation and bronchial responsiveness in interleukin-5 receptor alpha chain-deficient mice. Clin Exp Allergy. 2000. 30:874–881.85. Leckie MJ, ten Brinke A, Khan J, Diamant Z, O'Connor BJ, Walls CM, Mathur AK, Cowley HC, Chung KF, Djukanovic R, Hansel TT, Holgate ST, Sterk PJ, Barnes PJ. Effects of an interleukin-5 blocking monoclonal antibody on eosinophils, airway hyper-responsiveness, and the late asthmatic response. Lancet. 2000. 356:2144–2148.86. Flood-Page PT, Menzies-Gow AN, Kay AB, Robinson DS. Eosinophil's role remains uncertain as anti-interleukin-5 only partially depletes numbers in asthmatic airway. Am J Respir Crit Care Med. 2003. 167:199–204.87. Nair P, Pizzichini MM, Kjarsgaard M, Inman MD, Efthimiadis A, Pizzichini E, Hargreave FE, O'Byrne PM. Mepolizumab for prednisone-dependent asthma with sputum eosinophilia. N Engl J Med. 2009. 360:985–993.88. Haldar P, Brightling CE, Hargadon B, Gupta S, Monteiro W, Sousa A, Marshall RP, Bradding P, Green RH, Wardlaw AJ, Pavord ID. Mepolizumab and exacerbations of refractory eosinophilic asthma. N Engl J Med. 2009. 360:973–984.89. Pease JE. Asthma, allergy and chemokines. Curr Drug Targets. 2006. 7:3–12.90. Rothenberg ME, MacLean JA, Pearlman E, Luster AD, Leder P. Targeted disruption of the chemokine eotaxin partially reduces antigen-induced tissue eosinophilia. J Exp Med. 1997. 185:785–790.91. Matthews AN, Friend DS, Zimmermann N, Sarafi MN, Luster AD, Pearlman E, Wert SE, Rothenberg ME. Eotaxin is required for the baseline level of tissue eosinophils. Proc Natl Acad Sci U S A. 1998. 95:6273–6278.92. Yang Y, Loy J, Ryseck RP, Carrasco D, Bravo R. Antigen-induced eosinophilic lung inflammation develops in mice deficient in chemokine eotaxin. Blood. 1998. 92:3912–3923.93. Pope SM, Fulkerson PC, Blanchard C, Akei HS, Nikolaidis NM, Zimmermann N, Molkentin JD, Rothenberg ME. Identification of a cooperative mechanism involving interleukin-13 and eotaxin-2 in experimental allergic lung inflammation. J Biol Chem. 2005. 280:13952–13961.94. Pope SM, Zimmermann N, Stringer KF, Karow ML, Rothenberg ME. The eotaxin chemokines and CCR3 are fundamental regulators of allergen-induced pulmonary eosinophilia. J Immunol. 2005. 175:5341–5350.95. Pope SM, Brandt EB, Mishra A, Hogan SP, Zimmermann N, Matthaei KI, Foster PS, Rothenberg ME. IL-13 induces eosinophil recruitment into the lung by an IL-5- and eotaxin-dependent mechanism. J Allergy Clin Immunol. 2001. 108:594–601.96. Humbles AA, Lu B, Friend DS, Okinaga S, Lora J, Al-Garawi A, Martin TR, Gerard NP, Gerard C. The murine CCR3 receptor regulates both the role of eosinophils and mast cells in allergen-induced airway inflammation and hyperresponsiveness. Proc Natl Acad Sci U S A. 2002. 99:1479–1484.97. Ma W, Bryce PJ, Humbles AA, Laouini D, Yalcindag A, Alenius H, Friend DS, Oettgen HC, Gerard C, Geha RS. CCR3 is essential for skin eosinophilia and airway hyperresponsiveness in a murine model of allergic skin inflammation. J Clin Invest. 2002. 109:621–628.98. Justice JP, Borchers MT, Crosby JR, Hines EM, Shen HH, Ochkur SI, McGarry MP, Lee NA, Lee JJ. Ablation of eosinophils leads to a reduction of allergen-induced pulmonary pathology. Am J Physiol Lung Cell Mol Physiol. 2003. 284:L169–L178.99. Fulkerson PC, Fischetti CA, McBride ML, Hassman LM, Hogan SP, Rothenberg ME. A central regulatory role for eosinophils and the eotaxin/CCR3 axis in chronic experimental allergic airway inflammation. Proc Natl Acad Sci U S A. 2006. 103:16418–16423.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Eosinophils and childhood asthma
- Eosinophil cationic protein in relation to bronchial hyperresponsiveness in asthmatic children
- Serum eosinophil cationic protein(ECP) in children with atopic asthma
- The Efficacy of Eosinophilic Degranulation Proteins for Diagnosing Asthma in Chronic Cough Patients
- Eosinophil Apoptosis