Korean J Hematol.
2011 Dec;46(4):216-228. 10.5045/kjh.2011.46.4.216.
Human diversity of killer cell immunoglobulin-like receptors and disease
- Affiliations
-
- 1Department of Pathology and Laboratory Medicine, UCLA Immunogenetics Center, David Geffen School of Medicine at UCLA, University of California, Los Angeles, CA, USA. rrajalingam@mednet.ucla.edu
- KMID: 2251982
- DOI: http://doi.org/10.5045/kjh.2011.46.4.216
Abstract
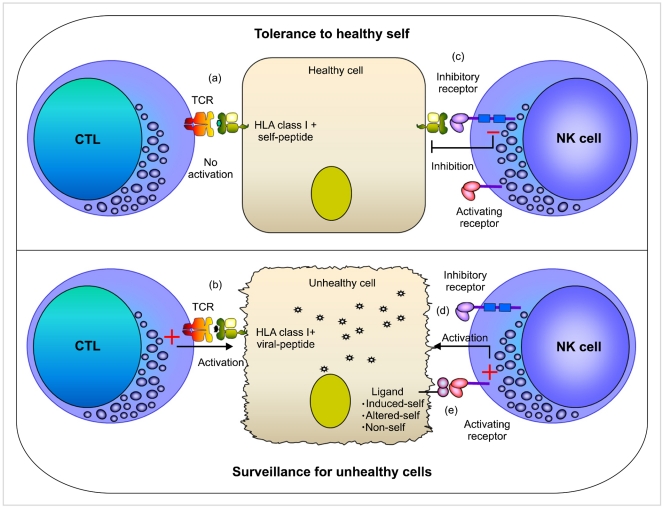
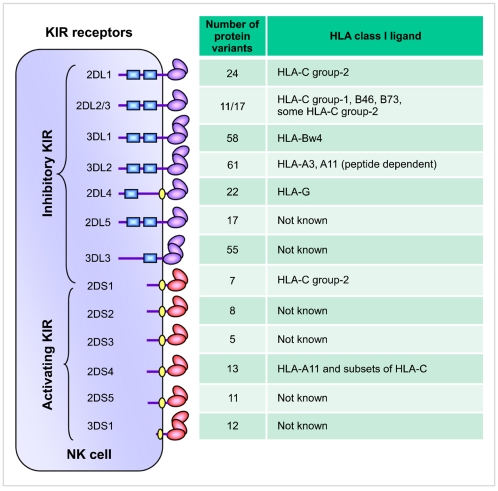
- Natural Killer (NK) cells are the third population of lymphocyte in the mononuclear cell compartment that triggers first-line of defense against viral infection and tumor transformation. Historically, NK cells were thought of as components of innate immunity based on their intrinsic ability to spontaneously kill target cells independent of HLA antigen restriction. However, it is now clear that NK cells are quite sophisticated and use a highly specific and complex target cell recognition receptor system arbitrated via a multitude of inhibitory and activating receptors. Killer cell immunoglobulin-like receptors (KIR) are the key receptors of human NK cells development and function. To date, fourteen distinct KIRs have been identified: eight are inhibitory types, and six are activating types. The number and type of KIR genes present varies substantially between individuals. Inhibitory KIRs recognize distinct motifs of polymorphic HLA class I molecules. Upon engagement of their specific HLA class I ligands, inhibitory KIR dampen NK cell reactivity. In contrast, activating KIRs are believed to stimulate NK cell reactivity when they sense their ligands (unknown). KIR and HLA gene families map to different human chromosomes (19 and 6, respectively), and their independent segregation produces a wide diversity in the number and type of inherited KIR-HLA combinations, likely contributing to overall immune competency. Consistent with this hypothesis, certain combinations of KIR-HLA variants have been correlated with susceptibility to diseases as diverse as autoimmunity, viral infections, and cancer. This review summarizes our emerging understanding of KIR-HLA diversity in human health and disease.
Keyword
MeSH Terms
Figure
Reference
-
1. Narni-Mancinelli E, Vivier E, Kerdiles YM. The 'T-cell-ness' of NK cells: unexpected similarities between NK cells and T cells. Int Immunol. 2011; 23:427–431. PMID: 21665959.
Article2. Kim S, Poursine-Laurent J, Truscott SM, et al. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature. 2005; 436:709–713. PMID: 16079848.
Article3. Sun JC, Beilke JN, Lanier LL. Immune memory redefined: characterizing the longevity of natural killer cells. Immunol Rev. 2010; 236:83–94. PMID: 20636810.
Article4. Bryceson YT, Long EO. Line of attack: NK cell specificity and integration of signals. Curr Opin Immunol. 2008; 20:344–352. PMID: 18439809.
Article5. Lanier LL. Natural killer cell receptor signaling. Curr Opin Immunol. 2003; 15:308–314. PMID: 12787756.
Article6. Valiante NM, Uhrberg M, Shilling HG, et al. Functionally and structurally distinct NK cell receptor repertoires in the peripheral blood of two human donors. Immunity. 1997; 7:739–751. PMID: 9430220.
Article8. McQueen KL, Parham P. Variable receptors controlling activation and inhibition of NK cells. Curr Opin Immunol. 2002; 14:615–621. PMID: 12183162.
Article9. Vivier E, Raulet DH, Moretta A, et al. Innate or adaptive immunity? The example of natural killer cells. Science. 2011; 331:44–49. PMID: 21212348.
Article10. Raulet DH, Vance RE, McMahon CW. Regulation of the natural killer cell receptor repertoire. Annu Rev Immunol. 2001; 19:291–330. PMID: 11244039.
Article11. Ljunggren HG, Karre K. In search of the 'missing self': MHC molecules and NK cell recognition. Immunol Today. 1990; 11:237–244. PMID: 2201309.
Article12. Abi-Rached L, Moesta AK, Rajalingam R, Guethlein LA, Parham P. Human-specific evolution and adaptation led to major qualitative differences in the variable receptors of human and chimpanzee natural killer cells. PLoS Genet. 2010; 6:e1001192. PMID: 21079681.
Article13. Wilson MJ, Torkar M, Haude A, et al. Plasticity in the organization and sequences of human KIR/ILT gene families. Proc Natl Acad Sci U S A. 2000; 97:4778–4783. PMID: 10781084.
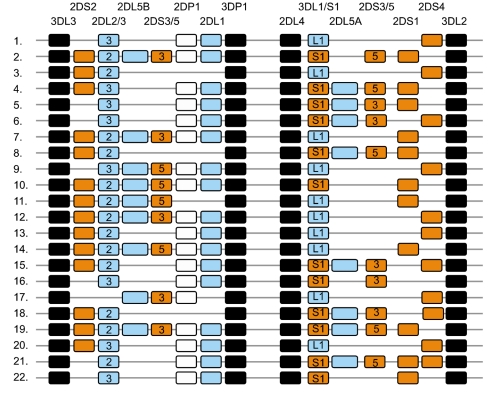
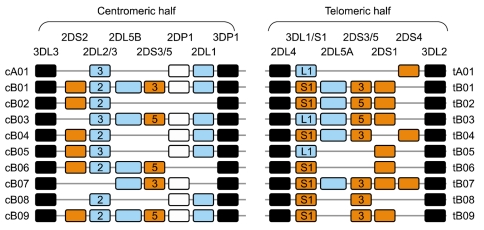
Article14. Uhrberg M, Parham P, Wernet P. Definition of gene content for nine common group B haplotypes of the Caucasoid population: KIR haplotypes contain between seven and eleven KIR genes. Immunogenetics. 2002; 54:221–229. PMID: 12136333.
Article15. Hsu KC, Liu XR, Selvakumar A, Mickelson E, O'Reilly RJ, Dupont B. Killer Ig-like receptor haplotype analysis by gene content: evidence for genomic diversity with a minimum of six basic framework haplotypes, each with multiple subsets. J Immunol. 2002; 169:5118–5129. PMID: 12391228.
Article16. Yawata M, Yawata N, Draghi M, Little AM, Partheniou F, Parham P. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J Exp Med. 2006; 203:633–645. PMID: 16533882.
Article17. Whang DH, Park H, Yoon JA, Park MH. Haplotype analysis of killer cell immunoglobulin-like receptor genes in 77 Korean families. Hum Immunol. 2005; 66:146–154. PMID: 15695000.
Article18. Middleton D, Meenagh A, Gourraud PA. KIR haplotype content at the allele level in 77 Northern Irish families. Immunogenetics. 2007; 59:145–158. PMID: 17200871.
Article19. Shilling HG, Guethlein LA, Cheng NW, et al. Allelic polymorphism synergizes with variable gene content to individualize human KIR genotype. J Immunol. 2002; 168:2307–2315. PMID: 11859120.
Article20. Garcia CA, Robinson J, Guethlein LA, Parham P, Madrigal JA, Marsh SG. Human KIR sequences 2003. Immunogenetics. 2003; 55:227–239. PMID: 12838379.
Article21. Winter CC, Gumperz JE, Parham P, Long EO, Wagtmann N. Direct binding and functional transfer of NK cell inhibitory receptors reveal novel patterns of HLA-C allotype recognition. J Immunol. 1998; 161:571–577. PMID: 9670929.22. Gardiner CM, Guethlein LA, Shilling HG, et al. Different NK cell surface phenotypes defined by the DX9 antibody are due to KIR3DL1 gene polymorphism. J Immunol. 2001; 166:2992–3001. PMID: 11207248.
Article23. Carr WH, Pando MJ, Parham P. KIR3DL1 polymorphisms that affect NK cell inhibition by HLA-Bw4 ligand. J Immunol. 2005; 175:5222–5229. PMID: 16210627.
Article24. Parham P, Norman PJ, Abi-Rached L, Guethlein LA. Variable NK cell receptors exemplified by human KIR3DL1/S1. J Immunol. 2011; 187:11–19. PMID: 21690332.
Article25. Norman PJ, Abi-Rached L, Gendzekhadze K, et al. Unusual selection on the KIR3DL1/S1 natural killer cell receptor in Africans. Nat Genet. 2007; 39:1092–1099. PMID: 17694054.
Article26. Vivian JP, Duncan RC, Berry R, et al. Killer cell immunoglobulin-like receptor 3DL1-mediated recognition of human leukocyte antigen B. Nature. 2011; 479:401–405. PMID: 22020283.
Article27. Hsu KC, Chida S, Geraghty DE, Dupont B. The killer cell immunoglobulin-like receptor (KIR) genomic region: gene-order, haplotypes and allelic polymorphism. Immunol Rev. 2002; 190:40–52. PMID: 12493005.
Article28. Yawata M, Yawata N, Abi-Rached L, Parham P. Variation within the human killer cell immunoglobulin-like receptor (KIR) gene family. Crit Rev Immunol. 2002; 22:463–482. PMID: 12803322.29. Norman PJ, Abi-Rached L, Gendzekhadze K, et al. Meiotic recombination generates rich diversity in NK cell receptor genes, alleles, and haplotypes. Genome Res. 2009; 19:757–769. PMID: 19411600.
Article30. Ordonez D, Meenagh A, Gómez-Lozano N, Castanño J, Middleton D, Vilches C. Duplication, mutation and recombination of the human orphan gene KIR2DS3 contribute to the diversity of KIR haplotypes. genes Immun. 2008; 9:431–437. PMID: 18480828.
Article31. Ashouri E, Farjadian S, Reed EF, Ghaderi A, Rajalingam R. KIR gene content diversity in four Iranian populations. Immunogenetics. 2009; 61:483–492. PMID: 19521696.
Article32. Uhrberg M, Valiante NM, Shum BP, et al. Human diversity in killer cell inhibitory receptor genes. Immunity. 1997; 7:753–763. PMID: 9430221.
Article33. Jiang K, Zhu FM, Lv QF, Yan LX. Distribution of killer cell immunoglobulin-like receptor genes in the Chinese Han population. Tissue Antigens. 2005; 65:556–563. PMID: 15896204.
Article34. Yawata M, Yawata N, McQueen KL, et al. Predominance of group A KIR haplotypes in Japanese associated with diverse NK cell repertoires of KIR expression. Immunogenetics. 2002; 54:543–550. PMID: 12439616.
Article35. Gendzekhadze K, Norman PJ, Abi-Rached L, Layrisse Z, Parham P. High KIR diversity in Amerindians is maintained using few gene-content haplotypes. Immunogenetics. 2006; 58:474–480. PMID: 16738943.
Article36. Ewerton PD, Leite Mde M, Magalhães M, Sena L, Melo dos Santos EJ. Amazonian Amerindians exhibit high variability of KIR profiles. Immunogenetics. 2007; 59:625–630. PMID: 17551723.
Article37. Toneva M, Lepage V, Lafay G, et al. Genomic diversity of natural killer cell receptor genes in three populations. Tissue Antigens. 2001; 57:358–362. PMID: 11380947.
Article38. Rajalingam R, Du Z, Meenagh A, et al. Distinct diversity of KIR genes in three southern Indian populations: comparison with world populations revealed a link between KIR gene content and pre-historic human migrations. Immunogenetics. 2008; 60:207–217. PMID: 18369612.
Article39. Rajalingam R, Krausa P, Shilling HG, et al. Distinctive KIR and HLA diversity in a panel of north Indian Hindus. Immunogenetics. 2002; 53:1009–1019. PMID: 11904677.
Article40. Kulkarni S, Single RM, Martin MP, et al. Comparison of the rapidly evolving KIR locus in Parsis and natives of India. Immunogenetics. 2008; 60:121–129. PMID: 18351333.
Article41. Marsh SG, Albert ED, Bodmer WF, et al. Nomenclature for factors of the HLA system, 2010. Tissue Antigens. 2010; 75:291–455. PMID: 20356336.42. Klein J, Sato A. The HLA system. Second of two parts. N Engl J Med. 2000; 343:782–786. PMID: 10984567.
Article43. Klein J, Sato A. The HLA system. First of two parts. N Engl J Med. 2000; 343:702–709. PMID: 10974135.44. Colonna M, Borsellino G, Falco M, Ferrara GB, Strominger JL. HLA-C is the inhibitory ligand that determines dominant resistance to lysis by NK1- and NK2-specific natural killer cells. Proc Natl Acad Sci U S A. 1993; 90:12000–12004. PMID: 8265660.
Article45. Wagtmann N, Biassoni R, Cantoni C, et al. Molecular clones of the p58 NK cell receptor reveal immunoglobulin-related molecules with diversity in both the extra- and intracellular domains. Immunity. 1995; 2:439–449. PMID: 7749980.
Article46. Winter CC, Long EO. A single amino acid in the p58 killer cell inhibitory receptor controls the ability of natural killer cells to discriminate between the two groups of HLA-C allotypes. J Immunol. 1997; 158:4026–4028. PMID: 9126959.47. Moesta AK, Norman PJ, Yawata M, Yawata N, Gleimer M, Parham P. Synergistic polymorphism at two positions distal to the ligand-binding site makes KIR2DL2 a stronger receptor for HLA-C than KIR2DL3. J Immunol. 2008; 180:3969–3979. PMID: 18322206.
Article48. Stewart CA, Laugier-Anfossi F, Vély F, et al. Recognition of peptide-MHC class I complexes by activating killer immunoglobulin-like receptors. Proc Natl Acad Sci U S A. 2005; 102:13224–13229. PMID: 16141329.
Article49. Graef T, Moesta AK, Norman PJ, et al. KIR2DS4 is a product of gene conversion with KIR3DL2 that introduced specificity for HLA-A*11 while diminishing avidity for HLA-C. J Exp Med. 2009; 206:2557–2572. PMID: 19858347.
Article50. Gumperz JE, Litwin V, Phillips JH, Lanier LL, Parham P. The Bw4 public epitope of HLA-B molecules confers reactivity with natural killer cell clones that express NKB1, a putative HLA receptor. J Exp Med. 1995; 181:1133–1144. PMID: 7532677.
Article51. Cella M, Longo A, Ferrara GB, Strominger JL, Colonna M. NK3-specific natural killer cells are selectively inhibited by Bw4-positive HLA alleles with isoleucine 80. J Exp Med. 1994; 180:1235–1242. PMID: 7931060.
Article52. Thananchai H, Gillespie G, Martin MP, et al. Cutting edge: allele-specific and peptide-dependent interactions between KIR3DL1 and HLA-A and HLA-B. J Immunol. 2007; 178:33–37. PMID: 17182537.
Article53. Pende D, Biassoni R, Cantoni C, et al. The natural killer cell receptor specific for HLA-A allotypes: a novel member of the p58/p70 family of inhibitory receptors that is characterized by three immunoglobulin-like domains and is expressed as a 140-kD disulphide-linked dimer. J Exp Med. 1996; 184:505–518. PMID: 8760804.
Article54. Hansasuta P, Dong T, Thananchai H, et al. Recognition of HLA-A3 and HLA-A11 by KIR3DL2 is peptide-specific. Eur J Immunol. 2004; 34:1673–1679. PMID: 15162437.
Article55. Rajagopalan S, Long EO. A human histocompatibility leukocyte antigen (HLA)-G-specific receptor expressed on all natural killer cells. J Exp Med. 1999; 189:1093–1100. PMID: 10190900.
Article56. Moffett-King A. Natural killer cells and pregnancy. Nat Rev Immunol. 2002; 2:656–663. PMID: 12209134.
Article57. Rajagopalan S, Bryceson YT, Kuppusamy SP, et al. Activation of NK cells by an endocytosed receptor for soluble HLA-G. PLoS Biol. 2006; 4:e9. PMID: 16366734.
Article58. Kikuchi-Maki A, Yusa S, Catina TL, Campbell KS. KIR2DL4 is an IL-2-regulated NK cell receptor that exhibits limited expression in humans but triggers strong IFN-gamma production. J Immunol. 2003; 171:3415–3425. PMID: 14500636.59. Faure M, Long EO. KIR2DL4 (CD158d), an NK cell-activating receptor with inhibitory potential. J Immunol. 2002; 168:6208–6214. PMID: 12055234.
Article60. Ponte M, Cantoni C, Biassoni R, et al. Inhibitory receptors sensing HLA-G1 molecules in pregnancy: decidua-associated natural killer cells express LIR-1 and CD94/NKG2A and acquire p49, an HLA-G1-specific receptor. Proc Natl Acad Sci U S A. 1999; 96:5674–5679. PMID: 10318943.
Article61. Martin MP, Gao X, Lee JH, et al. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet. 2002; 31:429–434. PMID: 12134147.
Article62. Du Z, Gjertson DW, Reed EF, Rajalingam R. Receptor-ligand analyses define minimal killer cell Ig-like receptor (KIR) in humans. Immunogenetics. 2007; 59:1–15. PMID: 17103212.
Article63. Single RM, Martin MP, Gao X, et al. Global diversity and evidence for coevolution of KIR and HLA. Nat Genet. 2007; 39:1114–1119. PMID: 17694058.
Article64. Gendzekhadze K, Norman PJ, Abi-Rached L, et al. Co-evolution of KIR2DL3 with HLA-C in a human population retaining minimal essential diversity of KIR and HLA class I ligands. Proc Natl Acad Sci U S A. 2009; 106:18692–18697. PMID: 19837691.
Article65. Yokoyama WM, Kim S. How do natural killer cells find self to achieve tolerance? Immunity. 2006; 24:249–257. PMID: 16546094.
Article66. Raulet DH, Vance RE. Self-tolerance of natural killer cells. Nat Rev Immunol. 2006; 6:520–531. PMID: 16799471.
Article67. Anfossi N, Andre P, Guia S, et al. Human NK cell education by inhibitory receptors for MHC class I. Immunity. 2006; 25:331–342. PMID: 16901727.
Article68. Carrington M, O'Brien SJ. The influence of HLA genotype on AIDS. Annu Rev Med. 2003; 54:535–551. PMID: 12525683.
Article69. Fellay J, Shianna KV, Ge D, et al. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007; 317:944–947. PMID: 17641165.
Article70. Goulder PJ, Brander C, Tang Y, et al. Evolution and transmission of stable CTL escape mutations in HIV infection. Nature. 2001; 412:334–338. PMID: 11460164.
Article71. Betts MR, Nason MC, West SM, et al. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006; 107:4781–4789. PMID: 16467198.
Article72. Qi Y, Martin MP, Gao X, et al. KIR/HLA pleiotropism: protection against both HIV and opportunistic infections. PLoS Pathog. 2006; 2:e79. PMID: 16933987.
Article73. Alter HJ, Seeff LB. Recovery, persistence, and sequelae in hepatitis C virus infection: a perspective on long-term outcome. Semin Liver Dis. 2000; 20:17–35. PMID: 10895429.
Article74. Khakoo SI, Thio CL, Martin MP, et al. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004; 305:872–874. PMID: 15297676.
Article75. Romero V, Azocar J, Zúñiga J, et al. Interaction of NK inhibitory receptor genes with HLA-C and MHC class II alleles in Hepatitis C virus infection outcome. Mol Immunol. 2008; 45:2429–2436. PMID: 18289678.
Article76. Lee IF, Qin H, Trudeau J, Dutz J, Tan R. Regulation of autoimmune diabetes by complete Freund's adjuvant is mediated by NK cells. J Immunol. 2004; 172:937–942. PMID: 14707066.
Article77. Fort MM, Leach MW, Rennick DM. A role for NK cells as regulators of CD4+ T cells in a transfer model of colitis. J Immunol. 1998; 161:3256–3261. PMID: 9759840.78. Xu W, Fazekas G, Hara H, Tabira T. Mechanism of natural killer (NK) cell regulatory role in experimental autoimmune encephalomyelitis. J Neuroimmunol. 2005; 163:24–30. PMID: 15885305.
Article79. Setiady YY, Pramoonjago P, Tung KS. Requirements of NK cells and proinflammatory cytokines in T cell-dependent neonatal autoimmune ovarian disease triggered by immune complex. J Immunol. 2004; 173:1051–1058. PMID: 15240693.
Article80. Shi FD, Wang HB, Li H, et al. Natural killer cells determine the outcome of B cell-mediated autoimmunity. Nat Immunol. 2000; 1:245–251. PMID: 10973283.
Article81. Suzuki Y, Hamamoto Y, Ogasawara Y, et al. Genetic polymorphisms of killer cell immunoglobulin-like receptors are associated with susceptibility to psoriasis vulgaris. J Invest Dermatol. 2004; 122:1133–1136. PMID: 15140215.
Article82. Holm SJ, Sakuraba K, Mallbris L, Wolk K, Stahle M, Sanchez FO. Distinct HLA-C/KIR genotype profile associates with guttate psoriasis. J Invest Dermatol. 2005; 125:721–730. PMID: 16185272.
Article83. Luszczek W, Mańczak M, Cisło M, et al. Gene for the activating natural killer cell receptor, KIR2DS1, is associated with susceptibility to psoriasis vulgaris. Hum Immunol. 2004; 65:758–766. PMID: 15310528.84. van der Slik AR, Koeleman BP, Verduijn W, Bruining GJ, Roep BO, Giphart MJ. KIR in type 1 diabetes: disparate distribution of activating and inhibitory natural killer cell receptors in patients versus HLA-matched control subjects. Diabetes. 2003; 52:2639–2642. PMID: 14514651.85. Momot T, Koch S, Hunzelmann N, et al. Association of killer cell immunoglobulin-like receptors with scleroderma. Arthritis Rheum. 2004; 50:1561–1565. PMID: 15146426.
Article86. Levinson RD, Du Z, Luo L, et al. Combination of KIR and HLA gene variants augments the risk of developing birdshot chorioretinopathy in HLA-A*29-positive individuals. Genes Immun. 2008; 9:249–258. PMID: 18340360.
Article87. Levinson RD, Okada AA, Ashouri E, Keino H, Rajalingam R. Killer cell immunoglobulin-like receptor gene-cluster 3DS1-2DL5-2DS1-2DS5 predisposes susceptibility to Vogt-Koyanagi-Harada syndrome in Japanese individuals. Hum Immunol. 2010; 71:192–194. PMID: 19897003.
Article88. Levinson RD, Martin TM, Luo L, et al. Killer Cell Immunoglobulin-like receptors in HLA-B27-associated acute anterior uveitis, with and without axial spondyloarthropathy. Invest Ophthalmol Vis Sci. 2010; 51:1505–1510. PMID: 19850842.
Article89. Waldhauer I, Steinle A. NK cells and cancer immunosurveillance. Oncogene. 2008; 27:5932–5943. PMID: 18836474.
Article90. Zamai L, Ponti C, Mirandola P, et al. NK cells and cancer. J Immunol. 2007; 178:4011–4016. PMID: 17371953.
Article91. Carrington M, Wang S, Martin MP, et al. Hierarchy of resistance to cervical neoplasia mediated by combinations of killer immunoglobulin-like receptor and human leukocyte antigen loci. J Exp Med. 2005; 201:1069–1075. PMID: 15809352.
Article92. Bonagura VR, Du Z, Ashouri E, et al. Activating killer cell immunoglobulin-like receptors 3DS1 and 2DS1 protect against developing the severe form of recurrent respiratory papillomatosis. Hum Immunol. 2010; 71:212–219. PMID: 19861144.
Article93. Kiessling R, Klein E, Pross H, Wigzell H. "Natural" killer cells in the mouse. II. Cytotoxic cells with specificity for mouse Moloney leukemia cells. Characteristics of the killer cell. Eur J Immunol. 1975; 5:117–121. PMID: 1086218.
Article94. Herberman RB, Nunn ME, Holden HT, Lavrin DH. Natural cytotoxic reactivity of mouse lymphoid cells against syngeneic and allogeneic tumors. II. Characterization of effector cells. Int J Cancer. 1975; 16:230–239. PMID: 1080480.
Article95. Valiante NM, Parham P. Natural killer cells, HLA class I molecules, and marrow transplantation. Biol Blood Marrow Transplant. 1997; 3:229–235. PMID: 9450917.96. Appelbaum FR. The current status of hematopoietic cell transplantation. Annu Rev Med. 2003; 54:491–512. PMID: 12414918.
Article97. Thomas ED. History, current results, and research in marrow transplantation. Perspect Biol Med. 1995; 38:230–237. PMID: 7899057.
Article98. Armitage JO. Bone marrow transplantation. N Engl J Med. 1994; 330:827–838. PMID: 8114836.
Article99. Shilling HG, Young N, Guethlein LA, et al. Genetic control of human NK cell repertoire. J Immunol. 2002; 169:239–247. PMID: 12077250.
Article100. Ruggeri L, Capanni M, Urbani E, et al. Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science. 2002; 295:2097–2100. PMID: 11896281.
Article101. Giebel S, Locatelli F, Lamparelli T, et al. Survival advantage with KIR ligand incompatibility in hematopoietic stem cell transplantation from unrelated donors. Blood. 2003; 102:814–819. PMID: 12689936.
Article102. Davies SM, Ruggieri L, DeFor T, et al. Evaluation of KIR ligand incompatibility in mismatched unrelated donor hematopoietic transplants. Killer immunoglobulin-like receptor. Blood. 2002; 100:3825–3827. PMID: 12393440.103. Kawase T, Matsuo K, Kashiwase K, et al. HLA mismatch combinations associated with decreased risk of relapse: implications for the molecular mechanism. Blood. 2009; 113:2851–2858. PMID: 18997170.
Article104. Clausen J, Wolf D, Petzer AL, et al. Impact of natural killer cell dose and donor killer-cell immunoglobulin-like receptor (KIR) genotype on outcome following human leucocyte antigen-identical haematopoietic stem cell transplantation. Clin Exp Immunol. 2007; 148:520–528. PMID: 17493020.
Article105. Willemze R, Rodrigues CA, Labopin M, et al. KIR-ligand incompatibility in the graft-versus-host direction improves outcomes after umbilical cord blood transplantation for acute leukemia. Leukemia. 2009; 23:492–500. PMID: 19151783.
Article106. Pende D, Marcenaro S, Falco M, et al. Anti-leukemia activity of alloreactive NK cells in KIR ligand-mismatched haploidentical HSCT for pediatric patients: evaluation of the functional role of activating KIR and redefinition of inhibitory KIR specificity. Blood. 2009; 113:3119–3129. PMID: 18945967.
Article107. Lowe EJ, Turner V, Handgretinger R, et al. T-cell alloreactivity dominates natural killer cell alloreactivity in minimally T-cell-depleted HLA-non-identical paediatric bone marrow transplantation. Br J Haematol. 2003; 123:323–326. PMID: 14531915.
Article108. Farag SS, Bacigalupo A, Eapen M, et al. The effect of KIR ligand incompatibility on the outcome of unrelated donor transplantation: a report from the center for international blood and marrow transplant research, the European blood and marrow transplant registry, and the Dutch registry. Biol Blood Marrow Transplant. 2006; 12:876–884. PMID: 16864058.
Article109. Cooley S, Weisdorf DJ, Guethlein LA, et al. Donor selection for natural killer cell receptor genes leads to superior survival after unrelated transplantation for acute myelogenous leukemia. Blood. 2010; 116:2411–2419. PMID: 20581313.
Article110. Venstrom JM, Gooley TA, Spellman S, et al. Donor activating KIR3DS1 is associated with decreased acute GVHD in unrelated allogeneic hematopoietic stem cell transplantation. Blood. 2010; 115:3162–3165. PMID: 20124216.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Human CD8+ T-Cell Populations That Express Natural Killer Receptors
- Killer Cell Immunoglobulin-like Receptor (KIR) Analysis in Adult Korean Patients with Acute Myeloid Leukemia
- HLA Mismatched Allogeneic Hematopoietic Stem Cell Transplantation
- Maternal killer-cell immunoglobulin-like receptors and paternal human leukocyte antigen ligands in recurrent pregnancy loss cases in Turkey
- A Case of Cytomegalovirus Pneumonia in a Healthy Infant