J Clin Neurol.
2015 Oct;11(4):311-318. 10.3988/jcn.2015.11.4.311.
Susceptibility Genes for Multiple Sclerosis Identified in a Gene-Based Genome-Wide Association Study
- Affiliations
-
- 1Center for Genetic Epidemiology and Genomics, School of Public Health, Soochow University, Suzhou, Jiangsu, People's Republic of China. leisf@suda.edu.cn
- 2Jiangsu Key Laboratory of Preventive and Translational Medicine for Geriatric Diseases, School of Public Health, Soochow University, Suzhou, Jiangsu, People's Republic of China.
- 3Department of Tuberculosis Control, Ningbo Municipal Center for Disease Control & Prevention, Ningbo, Zhejiang, People's Republic of China.
- KMID: 2179750
- DOI: http://doi.org/10.3988/jcn.2015.11.4.311
Abstract
- BACKGROUND AND PURPOSE
Multiple sclerosis (MS) is a demyelinating and inflammatory disease of the central nervous system. The aim of this study was to identify more genes associated with MS.
METHODS
Based on the publicly available data of the single-nucleotide polymorphism-based genome-wide association study (GWAS) from the database of Genotypes and Phenotypes, we conducted a powerful gene-based GWAS in an initial sample with 931 family trios, and a replication study sample with 978 cases and 883 controls. For interesting genes, gene expression in MS-related cells between MS cases and controls was examined by using publicly available datasets.
RESULTS
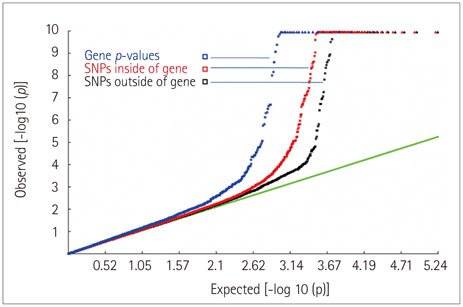
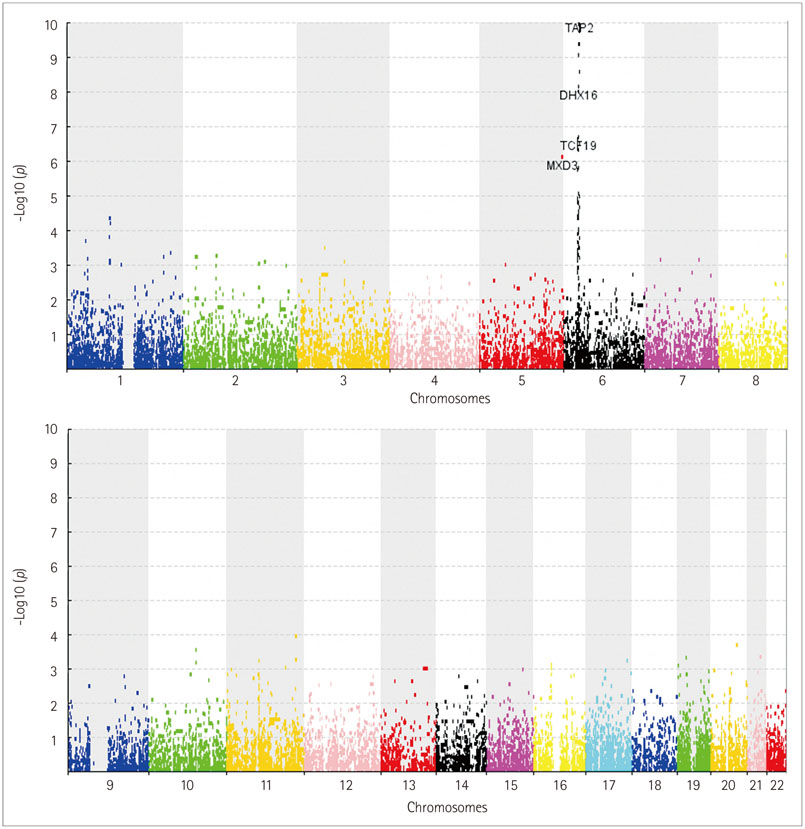
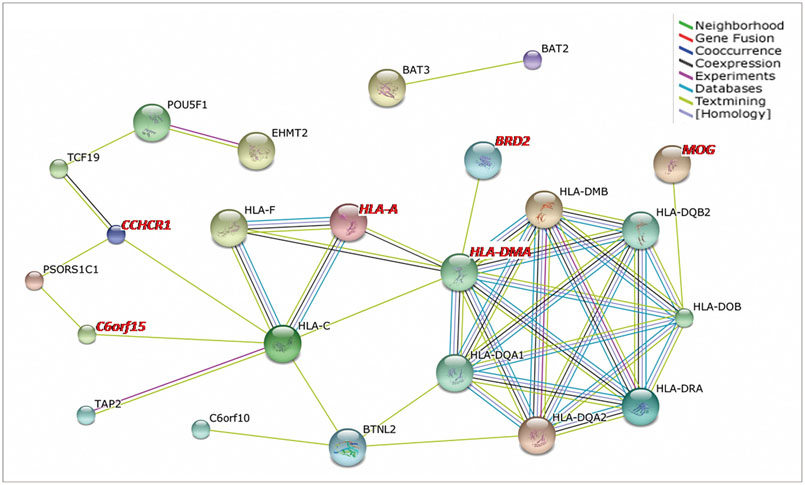
A total of 58 genes was identified, including 20 "novel" genes significantly associated with MS (p<1.40x10(-4)). In the replication study, 44 of the 58 identified genes had been genotyped and 35 replicated the association. In the gene-expression study, 21 of the 58 identified genes exhibited differential expressions in MS-related cells. Thus, 15 novel genes were supported by replicated association and/or differential expression. In particular, four of the novel genes, those encoding myelin oligodendrocyte glycoprotein (MOG), coiled-coil alpha-helical rod protein 1 (CCHCR1), human leukocyte antigen complex group 22 (HCG22), and major histocompatibility complex, class II, DM alpha (HLA-DMA), were supported by the evidence of both.
CONCLUSIONS
The results of this study emphasize the high power of gene-based GWAS in detecting the susceptibility genes of MS. The novel genes identified herein may provide new insights into the molecular genetic mechanisms underlying MS.
MeSH Terms
Figure
Reference
-
1. Selchen D, Bhan V, Blevins G, Devonshire V, Duquette P, Grand'Maison F, et al. MS, MRI, and the 2010 McDonald criteria: a Canadian expert commentary. Neurology. 2012; 79:23 Suppl 2. S1–S15.
Article2. Compston A, Coles A. Multiple sclerosis. Lancet. 2008; 372:1502–1517.
Article3. Milo R, Kahana E. Multiple sclerosis: geoepidemiology, genetics and the environment. Autoimmun Rev. 2010; 9:A387–A394.
Article4. Nakahara J, Maeda M, Aiso S, Suzuki N. Current concepts in multiple sclerosis: autoimmunity versus oligodendrogliopathy. Clin Rev Allergy Immunol. 2012; 42:26–34.
Article5. Australia and New Zealand Multiple Sclerosis Genetics Consortium (ANZgene). Genome-wide association study identifies new multiple sclerosis susceptibility loci on chromosomes 12 and 20. Nat Genet. 2009; 41:824–828.6. Jakkula E, Leppä V, Sulonen AM, Varilo T, Kallio S, Kemppinen A, et al. Genome-wide association study in a high-risk isolate for multiple sclerosis reveals associated variants in STAT3 gene. Am J Hum Genet. 2010; 86:285–291.
Article7. D'Netto MJ, Ward H, Morrison KM, Ramagopalan SV, Dyment DA, DeLuca GC, et al. Risk alleles for multiple sclerosis in multiplex families. Neurology. 2009; 72:1984–1988.8. International Multiple Sclerosis Genetics Consortium. Hafler DA, Compston A, Sawcer S, Lander ES, Daly MJ, et al. Risk alleles for multiple sclerosis identified by a genomewide study. N Engl J Med. 2007; 357:851–862.
Article9. Baranzini SE, Wang J, Gibson RA, Galwey N, Naegelin Y, Barkhof F, et al. Genome-wide association analysis of susceptibility and clinical phenotype in multiple sclerosis. Hum Mol Genet. 2009; 18:767–778.
Article10. Li MX, Gui HS, Kwan JS, Sham PC. GATES: a rapid and powerful gene-based association test using extended Simes procedure. Am J Hum Genet. 2011; 88:283–293.
Article11. Liu JZ, McRae AF, Nyholt DR, Medland SE, Wray NR, Brown KM, et al. A versatile gene-based test for genome-wide association studies. Am J Hum Genet. 2010; 87:139–145.
Article12. Neale BM, Sham PC. The future of association studies: gene-based analysis and replication. Am J Hum Genet. 2004; 75:353–362.
Article13. McDonald WI, Compston A, Edan G, Goodkin D, Hartung HP, Lublin FD, et al. Recommended diagnostic criteria for multiple sclerosis: guidelines from the International Panel on the diagnosis of multiple sclerosis. Ann Neurol. 2001; 50:121–127.
Article14. Kemppinen AK, Kaprio J, Palotie A, Saarela J. Systematic review of genome-wide expression studies in multiple sclerosis. BMJ Open. 2011; 1:e000053.
Article15. Lutterotti A, Jelcić I, Schulze C, Schippling S, Breiden P, Mazzanti B, et al. No proinflammatory signature in CD34+ hematopoietic progenitor cells in multiple sclerosis patients. Mult Scler. 2012; 18:1188–1192.
Article16. Annibali V, Ristori G, Angelini DF, Serafini B, Mechelli R, Cannoni S, et al. CD161(high)CD8+T cells bear pathogenetic potential in multiple sclerosis. Brain. 2011; 134(Pt 2):542–554.
Article17. Lieury A, Chanal M, Androdias G, Reynolds R, Cavagna S, Giraudon P, et al. Tissue remodeling in periplaque regions of multiple sclerosis spinal cord lesions. Glia. 2014; 62:1645–1658.
Article18. Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013; 41(Database issue):D808–D815.
Article19. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009; 4:44–57.
Article20. Graham RR, Kozyrev SV, Baechler EC, Reddy MV, Plenge RM, Bauer JW, et al. A common haplotype of interferon regulatory factor 5 (IRF5) regulates splicing and expression and is associated with increased risk of systemic lupus erythematosus. Nat Genet. 2006; 38:550–555.
Article21. Hochberg Y. A sharper Bonferroni procedure for multiple tests of significance. Biometrika. 1988; 75:800–802.
Article22. Hong MG, Reynolds CA, Feldman AL, Kallin M, Lambert JC, Amouyel P, et al. Genome-wide and gene-based association implicates FRMD6 in Alzheimer disease. Hum Mutat. 2012; 33:521–529.
Article23. Buil A, Martinez-Perez A, Perera-Lluna A, Rib L, Caminal P, Soria JM. A new gene-based association test for genome-wide association studies. BMC Proc. 2009; 3:Suppl 7. S130.
Article24. Patsopoulos NA, et al. Bayer Pharma MS Genetics Working Group. Steering Committees of Studies Evaluating IFNβ-1b and a CCR1-Antagonist. ANZgene Consortium. GeneMSA. International Multiple Sclerosis Genetic Consortium. Genome-wide meta-analysis identifies novel multiple sclerosis susceptibility loci. Ann Neurol. 2011; 70:897–912.
Article25. Weber F, Cepok S, Wolf C, Berthele A, Uhr M, Bettecken T, et al. Single-nucleotide polymorphisms in HLA- and non-HLA genes associated with the development of antibodies to interferon-β therapy in multiple sclerosis patients. Pharmacogenomics J. 2012; 12:238–245.
Article26. International Multiple Sclerosis Genetics Consortium. Wellcome Trust Case Control Consortium 2. Sawcer S, Hellenthal G, Pirinen M, Spencer CC, et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011; 476:214–219.
Article27. Song GG, Bae SC, Lee YH. Pathway analysis of genome-wide association studies on rheumatoid arthritis. Clin Exp Rheumatol. 2013; 31:566–574.28. Tervaniemi MH, Siitonen HA, Söderhäll C, Minhas G, Vuola J, Tiala I, et al. Centrosomal localization of the psoriasis candidate gene product, CCHCR1, supports a role in cytoskeletal organization. PLoS One. 2012; 7:e49920.
Article29. Gandhi G, Buttar BS, Albert L, Hasan Q, Aggarwal RK. Psoriasis-associated genetic polymorphism in North Indian population in the CCHCR1 gene and in a genomic segment flanking the HLA-C region. Dis Markers. 2011; 31:361–370.
Article30. Berger T, Reindl M. Multiple sclerosis: disease biomarkers as indicated by pathophysiology. J Neurol Sci. 2007; 259:21–26.
Article31. Wallström E, Khademi M, Andersson M, Weissert R, Linington C, Olsson T. Increased reactivity to myelin oligodendrocyte glycoprotein peptides and epitope mapping in HLA DR2(15)+ multiple sclerosis. Eur J Immunol. 1998; 28:3329–3335.
Article32. Raddassi K, Kent SC, Yang J, Bourcier K, Bradshaw EM, Seyfert-Margolis V, et al. Increased frequencies of myelin oligodendrocyte glycoprotein/MHC class II-binding CD4 cells in patients with multiple sclerosis. J Immunol. 2011; 187:1039–1046.
Article33. Van der Aa A, Hellings N, Bernard CC, Raus J, Stinissen P. Functional properties of myelin oligodendrocyte glycoprotein-reactive T cells in multiple sclerosis patients and controls. J Neuroimmunol. 2003; 137:164–176.
Article34. Linington C, Bradl M, Lassmann H, Brunner C, Vass K. Augmentation of demyelination in rat acute allergic encephalomyelitis by circulating mouse monoclonal antibodies directed against a myelin/oligodendrocyte glycoprotein. Am J Pathol. 1988; 130:443–454.35. Riker AI, Enkemann SA, Fodstad O, Liu S, Ren S, Morris C, et al. The gene expression profiles of primary and metastatic melanoma yields a transition point of tumor progression and metastasis. BMC Med Genomics. 2008; 1:13.
Article36. Zhang Z, Huettner PC, Nguyen L, Bidder M, Funk MC, Li J, et al. Aberrant promoter methylation and silencing of the POU2F3 gene in cervical cancer. Oncogene. 2006; 25:5436–5445.
Article37. Denis GV. Bromodomain coactivators in cancer, obesity, type 2 diabetes, and inflammation. Discov Med. 2010; 10:489–499.38. Filetici P, P O, Ballario P. The bromodomain: a chromatin browser? Front Biosci. 2001; 6:D866–D876.
Article39. Mahdi H, Fisher BA, Källberg H, Plant D, Malmström V, Rönnelid J, et al. Specific interaction between genotype, smoking and autoimmunity to citrullinated alpha-enolase in the etiology of rheumatoid arthritis. Nat Genet. 2009; 41:1319–1324.
Article40. Alcina A, Fernández O, Gonzalez JR, Catalá-Rabasa A, Fedetz M, Ndagire D, et al. Tag-SNP analysis of the GFI1-EVI5-RPL5-FAM69 risk locus for multiple sclerosis. Eur J Hum Genet. 2010; 18:827–831.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Genetic susceptibility factors of rheumatoid arthritis
- Sulwon Lecture 2009: The Search for Genetic Risk Factors of Type 2 Diabetes Mellitus
- Efficient Strategy to Identify Gene-Gene Interactions and Its Application to Type 2 Diabetes
- The Genetic Basis of Panic Disorder
- Relevance Epistasis Network of Gastritis for Intra-chromosomes in the Korea Associated Resource (KARE) Cohort Study