J Korean Med Sci.
2010 Apr;25(4):619-625. 10.3346/jkms.2010.25.4.619.
Expression Profiling of Calcium Induced Genes in Cultured Human Keratinocytes
- Affiliations
-
- 1Department of Dermatology and Research Institute for Medical Sciences, School of Medicine, Chungnam National University, Daejeon, Korea. jhoon@cnu.ac.kr
- 2Human Genomics Laboratory, Genome Research Center, Korea Research Institute of Bioscience and Biotechnology, Daejeon, Korea.
- 3Department of Dermatology, Sungkyunkwan University School of Medicine, Samsung Medical Center, Seoul, Korea.
- KMID: 1792935
- DOI: http://doi.org/10.3346/jkms.2010.25.4.619
Abstract
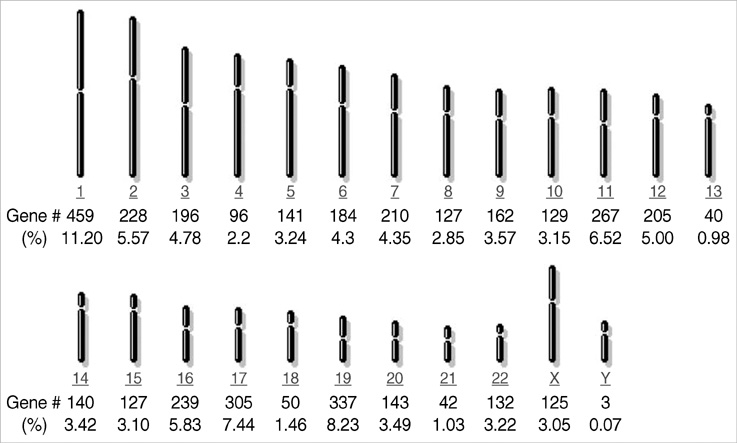
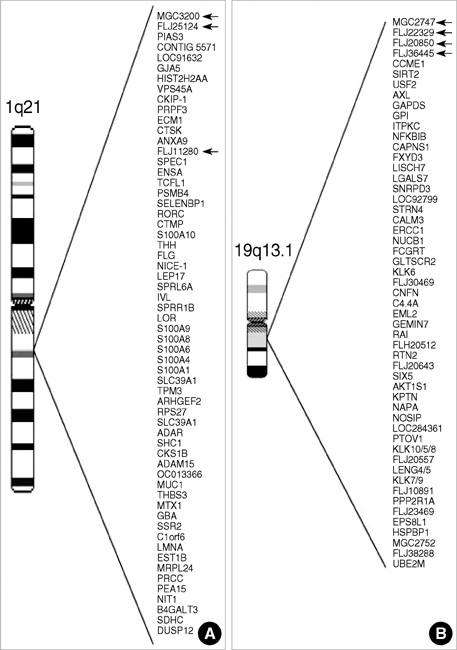
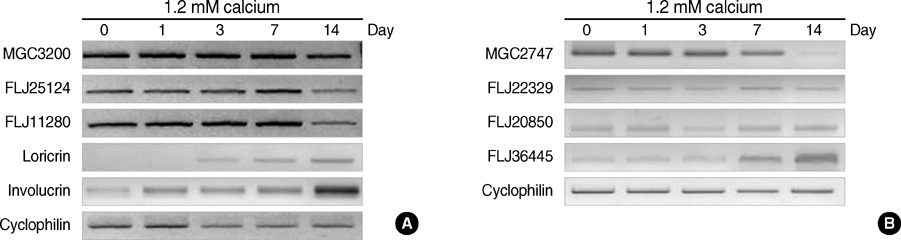
- Terminal differentiation of skin keratinocytes is a vertically directed multi-step process that is tightly controlled by the sequential expression of a variety of genes. To examine the gene expression profile in calcium-induced keratinocyte differentiation, we constructed a normalized cDNA library using mRNA isolated from these calcium-treated keratinocytes. After sequencing about 10,000 clones, we were able to obtain 4,104 independent genes. They consisted of 3,699 annotated genes and 405 expressed sequence tags (ESTs). Some were the genes involved in constituting epidermal structures and others were unknown genes that are probably associated with keratinocytes. In particular, we were able to identify genes located at the chromosome 1q21, the locus for the epidermal differentiation complex, and 19q13.1, another probable locus for epidermal differentiation-related gene clusters. One EST located at the chromosome 19q13.1 showed increased expression by calcium treatment, suggesting a novel candidate gene relevant to keratinocyte differentiation. These results demonstrate the complexity of the transcriptional profile of keratinocytes, providing important clues on which to base further investigations of the molecular events underlying keratinocyte differentiation.
MeSH Terms
Figure
Reference
-
1. Watt FM. Terminal differentiation of epidermal keratinocytes. Curr Opin Cell Biol. 1989. 1:1107–1115.
Article2. Chuong CM, Nickoloff BJ, Elias PM, Goldsmith LA, Macher E, Maderson PA, Sundberg JP, Tagami H, Plonka PM, Thestrup-Pederson K, Bernard BA, Schroder JM, Dotto P, Chang CM, Williams ML, Feingold KR, King LE, Kligman AM, Rees JL, Christophers E. What is the 'true' function of skin? Exp Dermatol. 2002. 11:159–187.3. Eckert RL, Crish JF, Robinson NA. The epidermal keratinocyte as a model for the study of gene regulation and cell differentiation. Physiol Rev. 1997. 77:397–424.
Article4. Seo EY, Namkung JH, Lee KM, Lee WH, Im M, Kee SH, Park GT, Yang JM, Seo YJ, Park JK, Kim CD, Lee JH. Analysis of calcium-inducible genes in keratinocytes using suppression subtractive hybridization and cDNA microarray. Genomics. 2005. 86:528–538.
Article5. Celis JE, Rasmussen HH, Gromov P, Olsen E, Madsen P, Leffers H, Honore B, Dejgaard K, Vorum H, Kristensen DB, Ostergaard M, Haunso A, Jensen NA, Celis A, Basse B, Lauridsen JB, Ratz GP, Andersen AH, Walbum E, Kjargaard I, Andersen I, Puype M, Damme JV, Vandekerckhove J. The human keratinocyte two-dimensional gel protein database (update 1995): mapping components of signal transduction pathways. Electrophoresis. 1995. 16:2177–2240.
Article6. Jansen BJ, van Ruissen F, de Jongh G, Zeeuwen PL, Schalkwijk J. Serial analysis of gene expression in differentiated cultures of human epidermal keratinocytes. J Invest Dermatol. 2001. 116:12–22.
Article7. van Ruissen F, Jansen BJ, de Jongh GJ, Zeeuwen PL, Schalkwijk J. A partial transcriptome of human epidermis. Genomics. 2002. 79:671–678.
Article8. Bonaldo MF, Lennon G, Soares MB. Normalization and subtraction: two approaches to facilitate gene discovery. Genome Res. 1996. 6:791–806.
Article9. Bonaldo MF, Yu MT, Jelenc P, Brown S, Su L, Lawton L, Deaven L, Efstratiadis A, Warburton D, Soares MB. Selection of cDNAs using chromosome-specific genomic clones: application to human chromosome 13. Hum Mol Genet. 1994. 3:1663–1673.
Article10. Soares MB, Bonaldo MF, Jelene P, Su L, Lawton L, Efstratiadis A. Construction and characterization of a normalized cDNA library. Proc Natl Acad Sci USA. 1994. 91:9228–9232.
Article11. Kim NS, Hahn Y, Oh JH, Lee JY, Oh KJ, Kim JM, Park HS, Kim S, Song KS, Rho SM, Yoo HS, Kim YS. Gene cataloging and expression profiling in human gastric cancer cells by expressed sequence tags. Genomics. 2004. 83:1024–1045.
Article12. Stanley JR, Yuspa SH. Specific epidermal protein markers are modulated during calcium-induced terminal differentiation. J Cell Biol. 1983. 96:1809–1814.
Article13. Bikle DD, Ratnam A, Mauro T, Harris J, Pillai S. Changes in calcium responsiveness and handling during keratinocyte differentiation. Potential role of the calcium receptor. J Clin Invest. 1996. 97:1085–1093.
Article14. Mammucari C, Tommasi di Vignano A, Sharov AA, Neilson J, Havrda MC, Roop DR, Botchkarev VA, Crabtree GR, Dotto GP. Integration of Notch 1 and calcineurin/NFAT signaling pathways in keratinocyte growth and differentiation control. Dev Cell. 2005. 8:665–676.
Article15. Grossi M, Hiou-Feige A, Tommasi Di Vignano A, Calautti E, Ostano P, Lee S, Chiorino G, Dotto GP. Negative control of keratinocyte differentiation by Rho/CRIK signaling coupled with up-regulation of KyoT1/2 (FHL1) expression. Proc Natl Acad Sci USA. 2005. 102:11313–11318.
Article16. Marenholz I, Zirra M, Fischer DF, Backendorf C, Ziegler A, Mischke D. Identification of human epidermal differentiation complex (EDC)-encoded genes by subtractive hybridization of entire YACs to a gridded keratinocyte cDNA library. Genome Res. 2001. 11:341–355.
Article17. Matsui T, Hayashi-Kisumi F, Kinoshita Y, Katahira S, Morita K, Miyachi Y, Ono Y, Imai T, Tanigawa Y, Komiya T, Tsukita S. Identification of novel keratinocyte-secreted peptides dermokine-alpha/-beta and a new stratified epithelium-secreted protein gene complex on human chromosome 19q13.1. Genomics. 2004. 84:384–397.18. Huang CM, Foster KW, DeSilva T, Zhang J, Shi Z, Yusuf N, Van Kampen KR, Elmets CA, Tang DC. Comparative proteomic profiling of murine skin. J Invest Dermatol. 2003. 121:51–64.
Article19. Lu J, Goldstein KM, Chen P, Huang S, Gelbert LM, Nagpal S. Transcriptional profiling of keratinocytes reveals a vitamin D-regulated epidermal differentiation network. J Invest Dermatol. 2005. 124:778–785.
Article20. Seo EY, Lee WH, Piao YJ, Kim KH, Lee KM, Ahn KS, Yang JM, Seo YJ, Kim CD, Park JK, Lee JH. Identification of calcium-inducible genes in primary keratinocytes using suppression-subtractive hybridization. Exp Dermatol. 2004. 13:163–169.
Article21. Lee WH, Jang S, Lee JS, Lee Y, Seo EY, You KH, Lee SC, Nam KI, Kim JM, Kee SH, Yang JM, Seo YJ, Park JK, Kim CD, Lee JH. Molecular cloning and expression of human keratinocyte proline-rich protein (hKPRP), an epidermal marker isolated from calcium-induced differentiating keratinocytes. J Invest Dermatol. 2005. 125:995–1000.
Article22. Crawford GE, Holt IE, Mullikin JC, Tai D, Blakesley R, Bouffard G, Young A, Masiello C, Green ED, Wolfsberg TG, Collins FS. National Institutes Of Health Intramural Sequencing Center. Identifying gene regulatory elements by genome-wide recovery of DNase hypersensitive sites. Proc Natl Acad Sci USA. 2004. 101:992–997.
Article23. Volz A, Korge BP, Compton JG, Ziegler A, Steinert PM, Mischke D. Physical mapping of a functional cluster of epidermal differentiation genes on chromosome 1q21. Genomics. 1993. 18:92–99.
Article24. Mischke D, Korge BP, Marenholz I, Volz A, Ziegler A. Genes encoding structural proteins of epidermal cornification and S100 calcium-binding proteins form a gene complex ("epidermal differentiation complex") on human chromosome 1q21. J Invest Dermatol. 1996. 106:989–992.
Article25. Oomizu S, Sahuc F, Asahina K, Inamatsu M, Matsuzaki T, Sasaki M, Obara M, Yoshizato K. Kdap, a novel gene associated with the stratification of the epithelium. Gene. 2000. 256:19–27.
Article26. Park GT, Lim SE, Jang SI, Morasso MI. Suprabasin, a novel epidermal differentiation marker and potential cornified envelope precursor. J Biol Chem. 2002. 277:45195–45202.
Article27. Elder JT, Zhao X. Evidence for local control of gene expression in the epidermal differentiation complex. Exp Dermatol. 2002. 11:406–412.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Expression of cytokeratins and involucrin in cultured human keratinocytes
- Expression of Toll-like Receptor 2 in Cultured Human Keratinocytes: The Effect of Bacterial Antigens, Cytokines and Calcium Concentration
- Effects of REtinoids on Keratinocytes HLA - DR and ICAM - 1 Expression Induced by Interferon - gamma
- Autocrine Extracellular Signal-regulated Kinase Activation in Normal Human Keratinocytes is not Interrupted by Calcium Triggering and is Involved in the Control of Cell Cycle at the Early Stage of Calciuminduced Differentiation
- Analysis of gene expression during mineralization of cultured human periodontal ligament cells