Ann Lab Med.
2013 May;33(3):159-166. 10.3343/alm.2013.33.3.159.
N-ras Mutation Detection by Pyrosequencing in Adult Patients with Acute Myeloid Leukemia at a Single Institution
- Affiliations
-
- 1Department of Laboratory Medicine, Gachon University Gil Medical Center, Incheon, Korea. jyahn@gilhospital.com
- 2Department of Medical Oncology and Hematology, Gachon University Gil Medical Center, Incheon, Korea.
- KMID: 1781320
- DOI: http://doi.org/10.3343/alm.2013.33.3.159
Abstract
- BACKGROUND
N-ras mutations are one of the most commonly detected abnormalities of myeloid origin. N-ras mutations result in a constitutively active N-ras protein that induces uncontrolled cell proliferation and inhibits apoptosis. We analyzed N-ras mutations in adult patients with AML at a particular institution and compared pyrosequencing analysis with a direct sequencing method for the detection of N-ras mutations.
METHODS
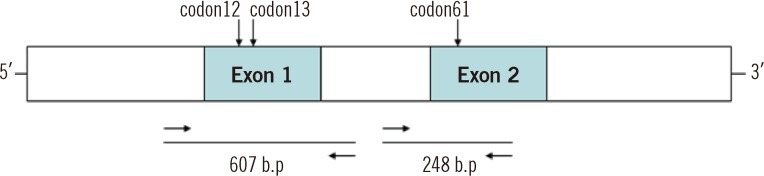
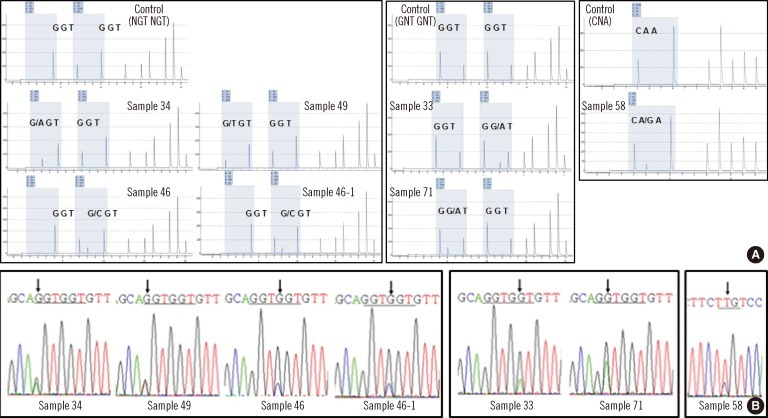
We analyzed 90 bone marrow samples from 83 AML patients. We detected N-ras mutations in codons 12, 13, and 61 using the pyrosequencing method and subsequently confirmed all data by direct sequencing. Using these methods, we screened the N-ras mutation quantitatively and determined the incidence and characteristic of N-ras mutation.
RESULTS
The incidence of N-ras mutation was 7.2% in adult AML patients. The patients with N-ras mutations showed significant higher hemoglobin levels (P=0.022) and an increased incidence of FLT3 mutations (P=0.003). We observed 3 cases with N-ras mutations in codon 12 (3.6%), 2 cases in codon 13 (2.4%), and 1 case in codon 61 (1.2%). All the mutations disappeared during chemotherapy.
CONCLUSIONS
There is a low incidence (7.2%) of N-ras mutations in AML patients compared with other populations. Similar data is obtained by both pyrosequencing and direct sequencing. This study showed the correlation between the N-ras mutation and the therapeutic response. However, pyrosequencing provides quantitative data and is useful for monitoring therapeutic responses.
Keyword
MeSH Terms
-
Adult
Aged
Aged, 80 and over
Antineoplastic Agents/therapeutic use
Bone Marrow/metabolism
Codon
Cytogenetic Analysis
Female
Hemoglobins/metabolism
Humans
Incidence
Leukemia, Myeloid, Acute/drug therapy/epidemiology/*genetics
Male
Middle Aged
Mutation
Sequence Analysis, DNA
fms-Like Tyrosine Kinase 3/genetics
ras Proteins/*genetics
Antineoplastic Agents
Codon
Hemoglobins
fms-Like Tyrosine Kinase 3
ras Proteins
Figure
Reference
-
1. Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nat Rev Cancer. 2003; 3:459–465. PMID: 12778136.
Article2. Karnoub AE, Weinberg RA. Ras oncogenes: split personalities. Nat Rev Mol Cell Biol. 2008; 9:517–531. PMID: 18568040.
Article3. Neubauer A, Dodge RK, George SL, Davey FR, Silver RT, Schiffer CA, et al. Prognostic importance of mutations in the ras proto-oncogenes in de novo acute myeloid leukemia. Blood. 1994; 83:1603–1611. PMID: 8123851.
Article4. De Melo MB, Lorand-Metze I, Lima CS, Saad ST, Costa FF. N-ras gene point mutations in Brazilian acute myelogenous leukemia patients correlate with a poor prognosis. Leuk Lymphoma. 1997; 24:309–317. PMID: 9156660.5. Bacher U, Haferlach T, Schoch C, Kern W, Schnittger S. Implications of NRAS mutations in AML: a study of 2502 patients. Blood. 2006; 107:3847–3853. PMID: 16434492.
Article6. Bowen DT, Frew ME, Hills R, Gale RE, Wheatley K, Groves MJ, et al. RAS mutation in acute myeloid leukemia is associated with distinct cytogenetic subgroups but does not influence outcome in patients younger than 60 years. Blood. 2005; 106:2113–2119. PMID: 15951308.
Article7. Ritter M, Kim TD, Lisske P, Thiede C, Schaich M, Neubauer A. Prognostic significance of N-RAS and K-RAS mutations in 232 patients with acute myeloid leukemia. Haematologica. 2004; 89:1397–1399. PMID: 15531466.8. Stirewalt DL, Kopecky KJ, Meshinchi S, Appelbaum FR, Slovak ML, Willman CL, et al. FLT3, RAS, and TP53 mutations in elderly patients with acute myeloid leukemia. Blood. 2001; 97:3589–3595. PMID: 11369655.
Article9. Schlenk RF, Döhner K, Krauter J, Fröhling S, Corbacioglu A, Bullinger L, et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008; 358:1909–1918. PMID: 18450602.
Article10. Ishikawa Y, Kiyoi H, Tsujimura A, Miyawaki S, Miyazaki Y, Kuriyama K, et al. Comprehensive analysis of cooperative gene mutations between class I and class II in de novo acute myeloid leukemia. Eur J Haematol. 2009; 83:90–98. PMID: 19309322.11. Patel JP, Gönen M, Figueroa ME, Fernandez H, Sun Z, Racevskis J, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012; 366:1079–1089. PMID: 22417203.
Article12. Rockova V, Abbas S, Wouters BJ, Erpelinck CA, Beverloo HB, Delwel R, et al. Risk stratification of intermediate-risk acute myeloid leukemia: integrative analysis of a multitude of gene mutation and gene expression markers. Blood. 2011; 118:1069–1076. PMID: 21596848.
Article13. Shen Y, Zhu YM, Fan X, Shi JY, Wang QR, Yan XJ, et al. Gene mutation patterns and their prognostic impact in a cohort of 1185 patients with acute myeloid leukemia. Blood. 2011; 118:5593–5603. PMID: 21881046.
Article14. Miyauchi J, Asada M, Sasaki M, Tsunematsu Y, Kojima S, Mizutani S, et al. Mutations of the N-ras gene in juvenile chronic myelogenous leukemia. Blood. 1994; 83:2248–2254. PMID: 8161790.
Article15. Ahmadian A, Ehn M, Hober S. Pyrosequencing: history, biochemistry and future. Clin Chim Acta. 2006; 363:83–94. PMID: 16165119.
Article16. Tsiatis AC, Norris-Kirby A, Rich RG, Hafez MJ, Gocke CD, Eshleman JR, et al. Comparison of Sanger sequencing, pyrosequencing, and melting curve analysis for the detection of KRAS mutations: diagnostic and clinical implications. J Mol Diagn. 2010; 12:425–432. PMID: 20431034.17. Ogino S, Kawasaki T, Brahmandam M, Yan L, Cantor M, Namgyal C, et al. Sensitive sequencing method for KRAS mutation detection by Pyrosequencing. J Mol Diagn. 2005; 7:413–421. PMID: 16049314.
Article18. Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, editors. WHO classification of tumours of haematopoietic and lymphoid tissues. 2008. 4th ed. Lyon: IARC;p. 110–155.19. Shaffer LG, Slovak ML, Campbell LJ, editors. ISCN 2009: An international system for cytogenetic nomenclature (2009). 2009. Basel: S Karger.20. Whitman SP, Ruppert AS, Radmacher MD, Mrózek K, Paschka P, Langer C, et al. FLT3 D835/I836 mutations are associated with poor disease-free survival and a distinct gene-expression signature among younger adults with de novo cytogenetically normal acute myeloid leukemia lacking FLT3 internal tandem duplications. Blood. 2008; 111:1552–1559. PMID: 17940205.
Article21. Illmer T, Thiede C, Fredersdorf A, Stadler S, Neubauer A, Ehninger G, et al. Activation of the RAS pathway is predictive for a chemosensitive phenotype of acute myelogenous leukemia blasts. Clin Cancer Res. 2005; 11:3217–3224. PMID: 15867216.
Article22. Darley RL, Burnett AK. Mutant RAS inhibits neutrophil but not macrophage differentiation and allows continued growth of neutrophil precursors. Exp Hematol. 1999; 27:1599–1608. PMID: 10560907.
Article23. Speletas M, Arvanitidi K, Tzoanopoulos D, Tsironidou V, Pardali E, Aggeli C, et al. Rapid mutational analysis of N-ras proto-oncogene in hematologic malignancies: study of 77 Greek patients. Haematologica. 2001; 86:918–927. PMID: 11532619.24. Auewarakul CU, Lauhakirti D, Tocharoentanaphol C. Frequency of RAS gene mutation and its cooperative genetic events in Southeast Asian adult acute myeloid leukemia. Eur J Haematol. 2006; 77:51–56. PMID: 16573741.
Article25. Tyner JW, Erickson H, Deininger MW, Willis SG, Eide CA, Levine RL, et al. High-throughput sequencing screen reveals novel, transforming RAS mutations in myeloid leukemia patients. Blood. 2009; 113:1749–1755. PMID: 19075190.
Article26. Chou FS, Wunderlich M, Griesinger A, Mulloy JC. N-Ras(G12D) induces features of stepwise transformation in preleukemic human umbilical cord blood cultures expressing the AML1-ETO fusion gene. Blood. 2011; 117:2237–2240. PMID: 21200020.
Article27. Sundström M, Edlund K, Lindell M, Glimelius B, Birgisson H, Micke P, et al. KRAS analysis in colorectal carcinoma: analytical aspects of Pyrosequencing and allele-specific PCR in clinical practice. BMC Cancer. 2010; 10:660. PMID: 21122130.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- A case of therapy-related acute myeloid leukemia with inv(16)(p13.1q22) after single low-dose iodine-131 treatment for thyroid cancer
- Mutation of the N-ras Gene in a Patient Suffering from the Blast Phase of Chronic Myelogenous Leukemia
- Acute Myeloid Leukemia with Intracardiac Thrombus Presenting as Acute Limb Ischemia
- Effects of Somatic Mutations Are Associated with SNP in the Progression of Individual Acute Myeloid Leukemia Patient: The Two-Hit Theory Explains Inherited Predisposition to Pathogenesis
- Myeloid Sarcoma of Peritoneum in Acute Myeloid Leukemia Patient with Inversion of Chromosome 16