J Bacteriol Virol.
2012 Jun;42(2):152-155. 10.4167/jbv.2012.42.2.152.
Molecular Typing of Clinical Cryptococcus gattii Isolates in Korea
- Affiliations
-
- 1Department of Clinical Laboratory Science, Catholic University of Pusan, Busan, Korea. smhwang@cup.ac.kr
- KMID: 1717685
- DOI: http://doi.org/10.4167/jbv.2012.42.2.152
Abstract
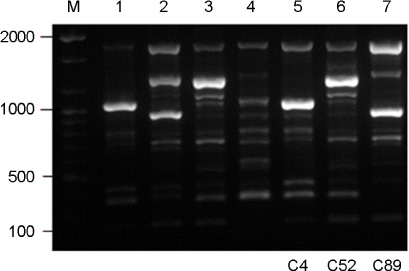
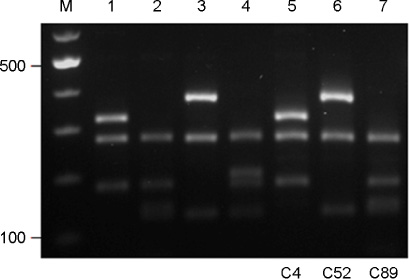
- Cryptococcus gattii causes life-threatening yeast infection in the pulmonary and central nervous systems of humans and animals, and traditionally has been considered to restrict into the tropical and subtropical areas. Despite rare incidence of cryptococcosis caused by C. gattii in Korea, three strains of C. gattii isolated from cryptococcosis patients between 1993 and 2010 were identified. To determine the genetic diversity, 3 strains of C. gattii were typed using PCR fingerprinting with primer M13 and the restriction fragment length polymorphism (RFLP) of orotidine monophosphosphate pyrophosphorylase (URA5) gene. All isolates were identified as serotype B and MATalpha mating type. The molecular types of each strain, on the other hand, turned out to be distinct belonging to VGI, VGII or III types, respectively. Although the travel histories of the patients were not available, clinical C. gattii strains isolated in Korea may represent the diverse molecular types existing worldwide.
MeSH Terms
Figure
Reference
-
1. Kwon-Chung KJ, Varma A. Do major species concepts support one, two or more species within Cryptococcus neoformans? FEMS Yeast Res. 2006. 6:574–587.
Article2. Bennett JE, Kwon-Chung KJ, Howard DH. Epidemiologic differences among serotypes of Cryptococcus neoformans. Am J Epidemiol. 1977. 105:582–586.3. Okamoto K, Hatakeyama S, Itoyama S, Nukui Y, Yoshino Y, Kitazawa T, et al. Cryptococcus gattii genotype VGIIa infection in Man, Japan, 2007. Emerg Infect Dis. 2010. 16:1155–1157.4. Kwon-Chung KJ, Bennett JE. Epidemiologic differences between the two varieties of Cryptococcus neoformans. Am J Epidemiol. 1984. 120:123–130.
Article5. Franzot SP, Salkin IF, Casadevall A. Cryptococcus neoformans var. grubii: separate varietal status for Cryptococcus neoformans serotype A isolates. J Clin Microbiol. 1999. 37:838–840.
Article6. Casadevall A, Perfect JR. Cryptococcus neoformans. 1998. Washington: ASM.7. Ellis DH. Cryptococcus neoformans var. gattii in Australia. J Clin Microbiol. 1987. 25:430–431.8. Dixit A, Carroll SF, Qureshi ST. Cryptococcus gattii: an emerging cause of fungal disease in North America. Interdiscip Perspect Infect Dis. 2009. 2009:840452.9. Kidd SE, Hagen F, Tscharke RL, Huynh M, Bartlett KH, Fyfe M, et al. A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada). Proc Natl Acad Sci U S A. 2004. 101:17258–17263.
Article10. Datta K, Bartlett KH, Marr KA. Cryptococcus gattii: emergence in western North America: exploitation of a novel ecological niche. Interdiscip Perspect Infect Dis. 2009. 2009:176532.11. Meyer W, Marszewska K, Amirmostofian M, Igreja RP, Hardtke C, Methllng K, et al. Molecular typing of global isolates of Cryptococcus neoformans var. neoformans by polymerase chain reaction fingerprinting and randomly amplified polymorphic DNA-a pilot study to standardize techniques on which to base a detailed epidemiological survey. Electrophoresis. 1999. 20:1790–1799.
Article12. Choi YH, Ngamskulrungroj P, Varma A, Sionov E, Hwang SM, Carriconde F, et al. Prevalence of the VNIc genotype of Cryptococcus neoformans in non-HIV-associated cryptococcosis in the Republic of Korea. FEMS Yeast Res. 2010. 10:769–778.
Article13. Springer DJ, Chaturvedi V. Projecting global occurrence of Cryptococcus gattii. Emerg Infect Dis. 2010. 16:14–20.14. Byrnes EJ 3rd, Li W, Ren P, Lewit Y, Voelz K, Fraser JA, et al. A diverse population of Cryptococcus gattii molecular type VGIII in southern California HIV/AIDS patients. PLoS Pathog. 2011. 7:e1002205.15. Kwon-Chung KJ, Polacheck I, Bennett JE. Improved diagnostic medium for separation of Cryptococcus neoformans var. neoformans (serotypes A and D) and Cryptococcus neoformans var. gattii (serotypes B and C). J Clin Microbiol. 1982. 15:535–537.
Article16. Meyer W, Castañeda A, Jackson S, Huynh M, Castañeda E. Molecular typing of Ibero American Cryptococcus neoformans isolates. Emerg Infect Dis. 2003. 9:189–195.17. Chen J, Varma A, Diaz MR, Litvintseva AP, Wollenberg KK, Kwon-Chung KJ. Cryptococcus neoformans strains and infection in apparently immunocompetent patients, China. Emerg Infect Dis. 2008. 14:755–762.
Article18. Hwang SM. Serotyping of Cryptococcus neoformans strains isolated in Korea. J Microbiol. 2002. 40:166–169.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Genotypes of Clinical and Environmental Isolates of Cryptococcus neoformans and Cryptococcus gattii in Korea
- Molecular Typing of Cryptococcus neoformans Isolated from Korean Patients
- Determining Potential Link between Environmental and Clinical Isolates of Cryptococcus neoformans/Cryptococcus gattii Species Complexes Using Phenotypic and Genotypic Characterisation
- Molecular Epidemiology of Clinical Cryptococcus neoformans Isolates in Seoul, Korea
- Differentiation of Varieties and Susceptibility Testing for Two Strains of Cryptococcus neoformans