Immune Netw.
2014 Feb;14(1):30-37. 10.4110/in.2014.14.1.30.
Co-stimulation of TLR4 and Dectin-1 Induces the Production of Inflammatory Cytokines but not TGF-beta for Th17 Cell Differentiation
- Affiliations
-
- 1Department of Microbiology and Immunology, The University of Michigan Medical School, Ann Arbor, MI 48109, USA. jchang24@mgh.harvard.edu
- 2Division of Gerontology, Department of Medicine, Beth Israel Deaconess Medical Center, Boston, MA 02215, USA.
- KMID: 1508826
- DOI: http://doi.org/10.4110/in.2014.14.1.30
Abstract
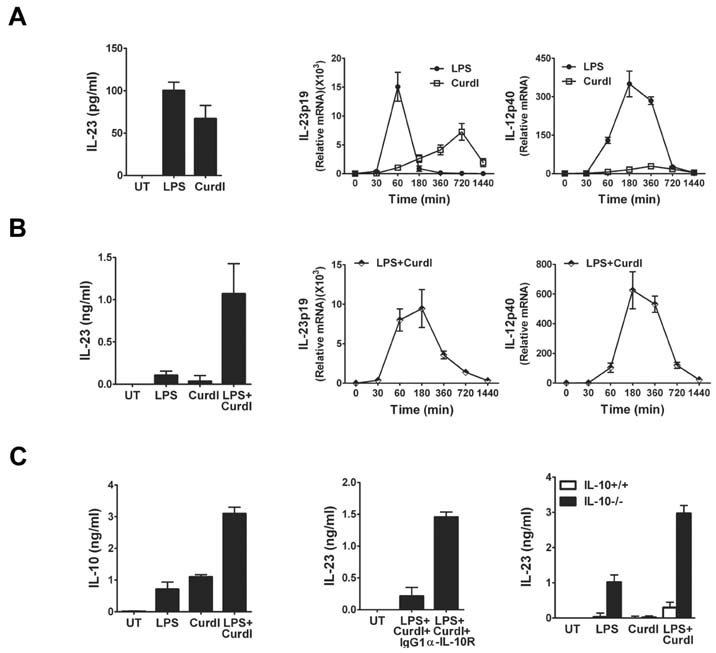
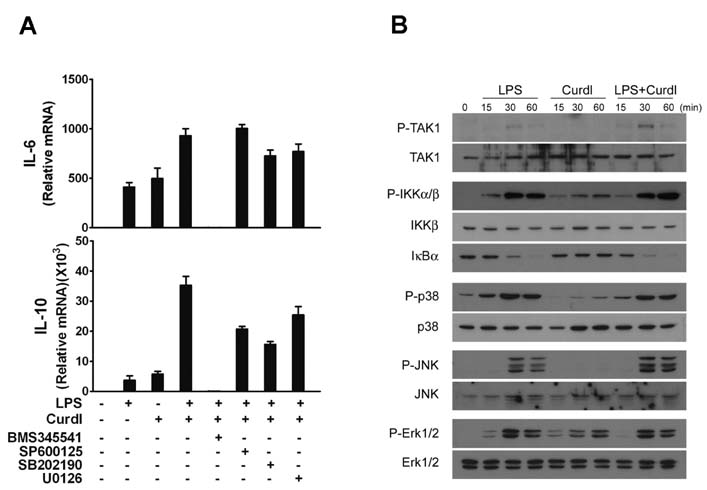
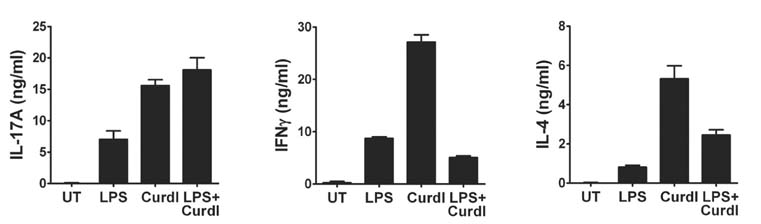
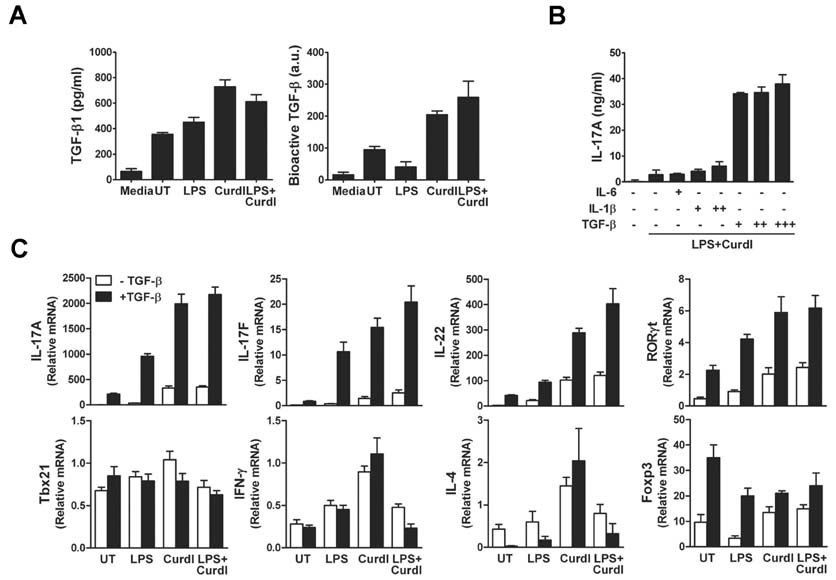
- Collaboration of TLR and non-TLR pathways in innate immune cells, which acts in concert for the induction of inflammatory cytokines, can mount a specific adaptive immune response tailored to a pathogen. Here, we show that murine DC produced increased IL-23 and IL-6 when they were treated with LPS together with curdlan that activates TLR4 and dectin-1, respectively. We also found that the induction of the inflammatory cytokine production by LPS and curdlan requires activation of IKK. However, the same treatment did not induce DC to produce a sufficient amount of TGF-beta. As a result, the conditioned media from DC treated with LPS and curdlan was not able to direct CD4+ T cells to Th17 cells. Addition of TGF-beta but not IL-6 or IL-1beta was able to promote IL-17 production from CD4+ T cells. Our results showed that although signaling mediated by LPS together with curdlan is a potent stimulator of DC to secrete many pro-inflammatory cytokines, TGF-beta production is a limiting factor for promoting Th17 immunity.
MeSH Terms
Figure
Reference
-
1. Palm NW, Medzhitov R. Pattern recognition receptors and control of adaptive immunity. Immunol Rev. 2009; 227:221–233.
Article2. Hollmig ST, Ariizumi K, Cruz PD Jr. Recognition of non-self-polysaccharides by C-type lectin receptors dectin-1 and dectin-2. Glycobiology. 2009; 19:568–575.
Article3. Brikos C, O'Neill LA. Signalling of toll-like receptors. Handb Exp Pharmacol. 2008; 21–50.
Article4. Taylor PR, Tsoni SV, Willment JA, Dennehy KM, Rosas M, Findon H, Haynes K, Steele C, Botto M, Gordon S, Brown GD. Dectin-1 is required for beta-glucan recognition and control of fungal infection. Nat Immunol. 2007; 8:31–38.
Article5. Saijo S, Fujikado N, Furuta T, Chung SH, Kotaki H, Seki K, Sudo K, Akira S, Adachi Y, Ohno N, Kinjo T, Nakamura K, Kawakami K, Iwakura Y. Dectin-1 is required for host defense against Pneumocystis carinii but not against Candida albicans. Nat Immunol. 2007; 8:39–46.
Article6. Carrion Sde J, Leal SM Jr, Ghannoum MA, Aimanianda V, Latge JP, Pearlman E. The RodA hydrophobin on Aspergillus fumigatus spores masks dectin-1- and dectin-2-dependent responses and enhances fungal survival in vivo. J Immunol. 2013; 191:2581–2588.7. LeibundGut-Landmann S, Gross O, Robinson MJ, Osorio F, Slack EC, Tsoni SV, Schweighoffer E, Tybulewicz V, Brown GD, Ruland J, Reis e Sousa C. Syk- and CARD9-dependent coupling of innate immunity to the induction of T helper cells that produce interleukin 17. Nat Immunol. 2007; 8:630–638.
Article8. O'Shea JJ, Steward-Tharp SM, Laurence A, Watford WT, Wei L, Adamson AS, Fan S. Signal transduction and Th17 cell differentiation. Microbes Infect. 2009; 11:599–611.9. Weaver CT, Harrington LE, Mangan PR, Gavrieli M, Murphy KM. Th17: an effector CD4 T cell line-age with regulatory T cell ties. Immunity. 2006; 24:677–688.
Article10. Zhou L, Ivanov II, Spolski R, Min R, Shenderov K, Egawa T, Levy DE, Leonard WJ, Littman DR. IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat Immunol. 2007; 8:967–974.
Article11. McGeachy MJ, Chen Y, Tato CM, Laurence A, Joyce-Shaikh B, Blumenschein WM, McClanahan TK, O'Shea JJ, Cua DJ. The interleukin 23 receptor is essential for the terminal differentiation of interleukin 17-producing effector T helper cells in vivo. Nat Immunol. 2009; 10:314–324.
Article12. Mangan PR, Harrington LE, O'Quinn DB, Helms WS, Bullard DC, Elson CO, Hatton RD, Wahl SM, Schoeb TR, Weaver CT. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 2006; 441:231–234.
Article13. Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 2006; 24:179–189.
Article14. Li MO, Wan YY, Sanjabi S, Robertson AK, Flavell RA. Transforming growth factor-beta regulation of immune responses. Annu Rev Immunol. 2006; 24:99–146.15. Oh SA, Li MO. TGF-beta: guardian of T cell function. J Immunol. 2013; 191:3973–3979.16. Yao Y, Li W, Kaplan MH, Chang CH. Interleukin (IL)-4 inhibits IL-10 to promote IL-12 production by dendritic cells. J Exp Med. 2005; 201:1899–1903.
Article17. Tesseur I, Zou K, Berber E, Zhang H, Wyss-Coray T. Highly sensitive and specific bioassay for measuring bioactive TGF-beta. BMC Cell Biol. 2006; 7:15.18. Chang J, Voorhees TJ, Liu Y, Zhao Y, Chang CH. Interleukin-23 production in dendritic cells is negatively regulated by protein phosphatase 2A. Proc Natl Acad Sci USA. 2010; 107:8340–8345.
Article19. Taylor AW. Review of the activation of TGF-beta in immunity. J Leukoc Biol. 2009; 85:29–33.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Dectin-1 Stimulation Selectively Reinforces LPS-driven IgG1 Production by Mouse B Cells
- The Production and Correlation of Silica Induced Proinflammatory Cytokines and TGF-beta from Monocytes of Balb/C Mice
- The Effect of Inflammatory Cytokines on the Differentiation of Th17 Cells in Human Peripheral Blood
- Targeting Interleukin-17 and Th17 in Immune Inflammatory Diseases
- Ciglitazone, a Peroxisome Proliferator-Activated Receptor Gamma Ligand, Inhibits Proliferation and Differentiation of Th17 Cells