Korean J Physiol Pharmacol.
2024 Sep;28(5):479-491. 10.4196/kjpp.2024.28.5.479.
Network pharmacology prediction to discover the potential pharmacological action mechanism of Rhizoma Dioscoreae for liver regeneration
- Affiliations
-
- 1Department of Hepatobiliary Surgery, The Second Affiliated Hospital, Dalian Medical University, Dalian 116021, Liaoning
- 2College of Basic Medical Sciences, Dalian Medical University, Dalian 116044, China
- 3Department of Traditional Chinese Medicine, The First Affiliated Hospital of Dalian Medical University, Dalian 116014, China
- KMID: 2559140
- DOI: http://doi.org/10.4196/kjpp.2024.28.5.479
Abstract
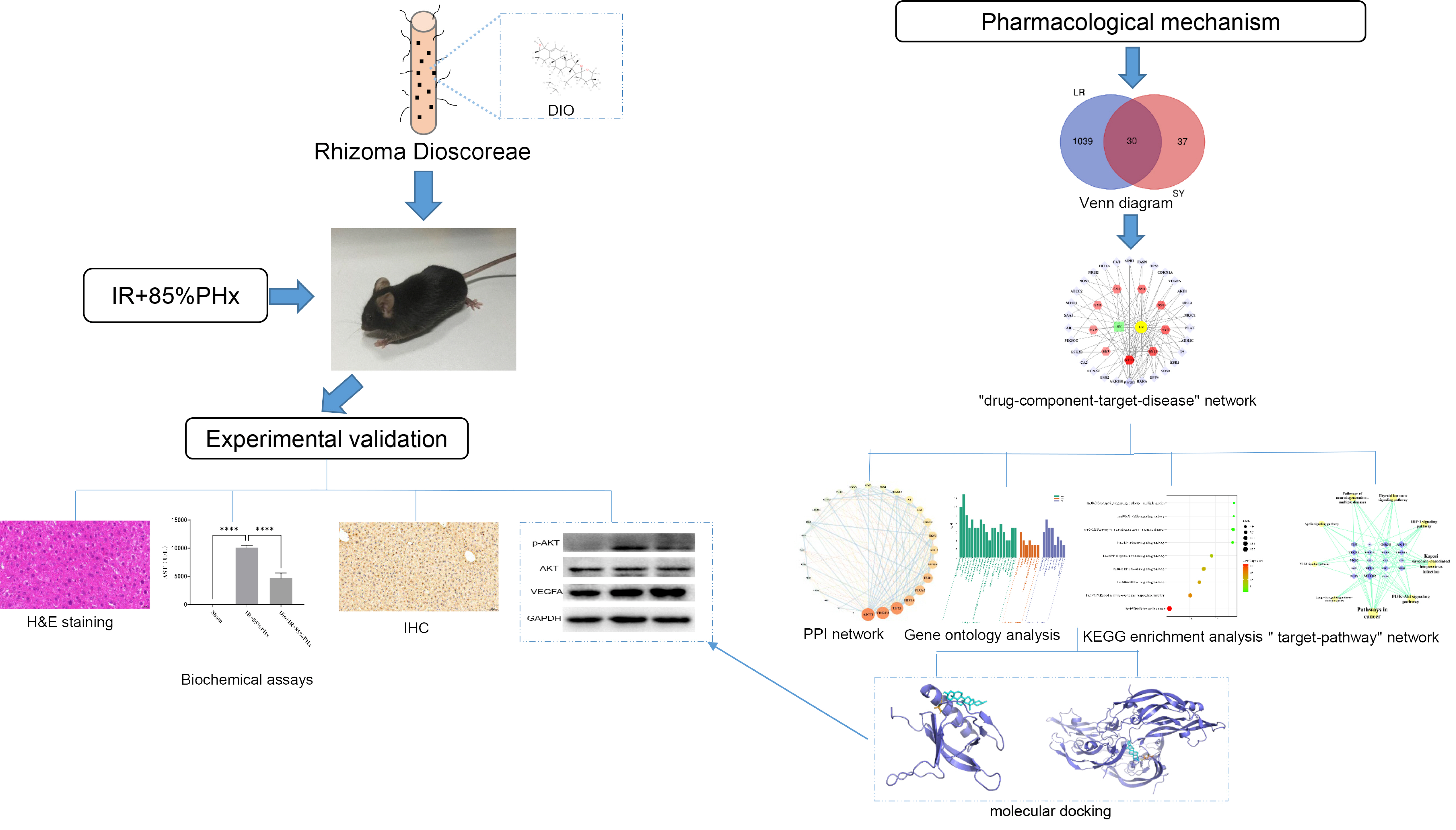
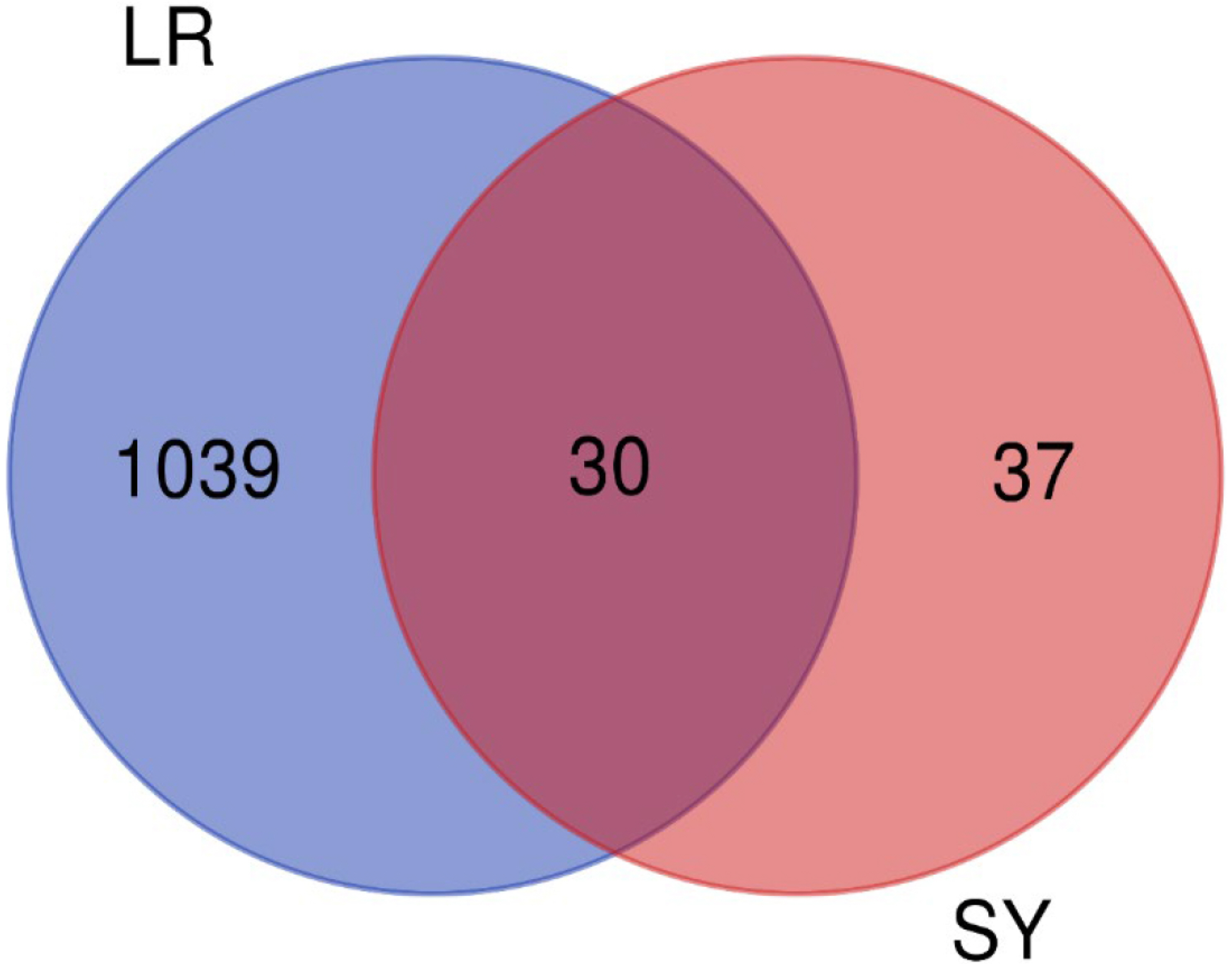
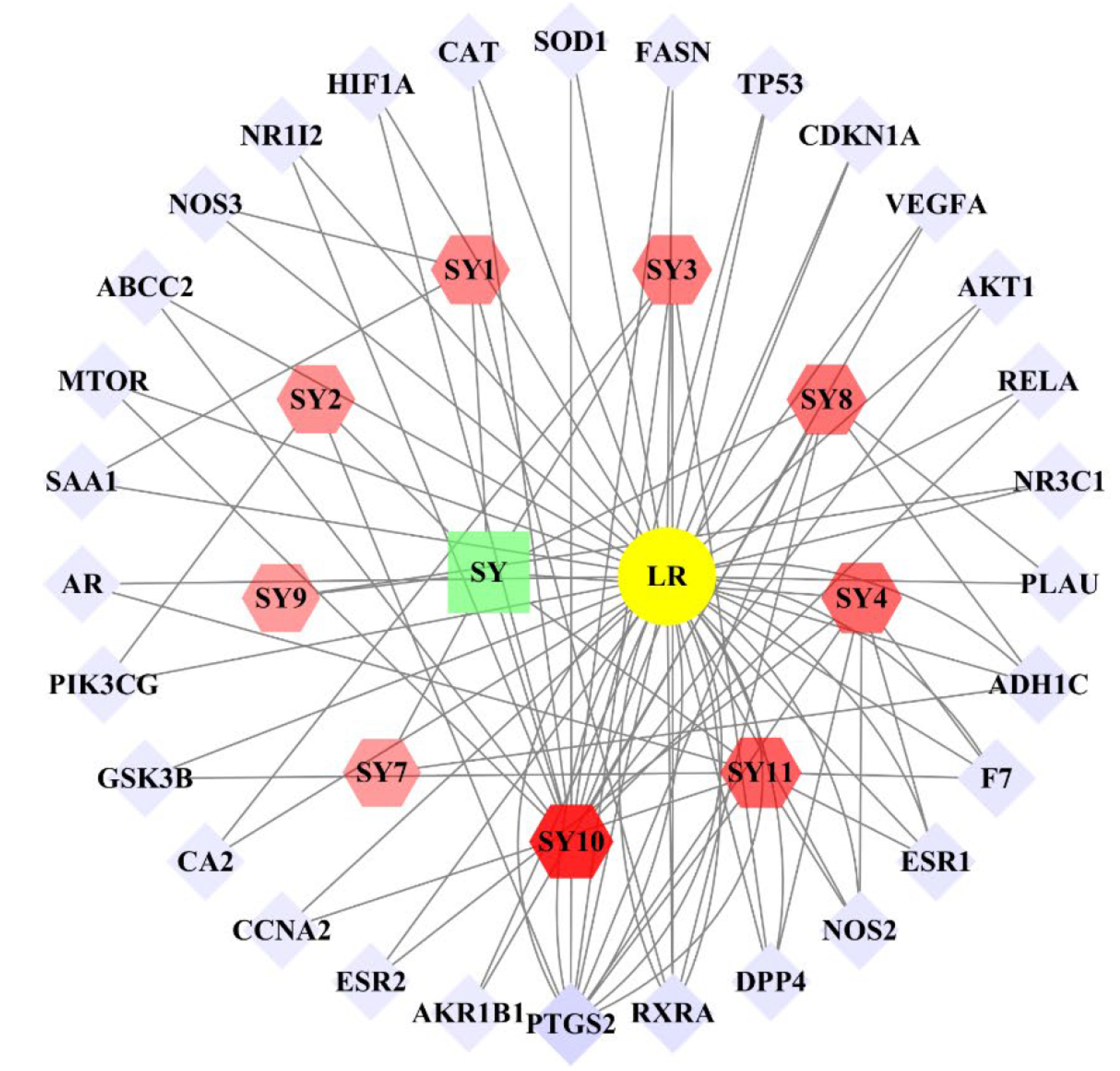
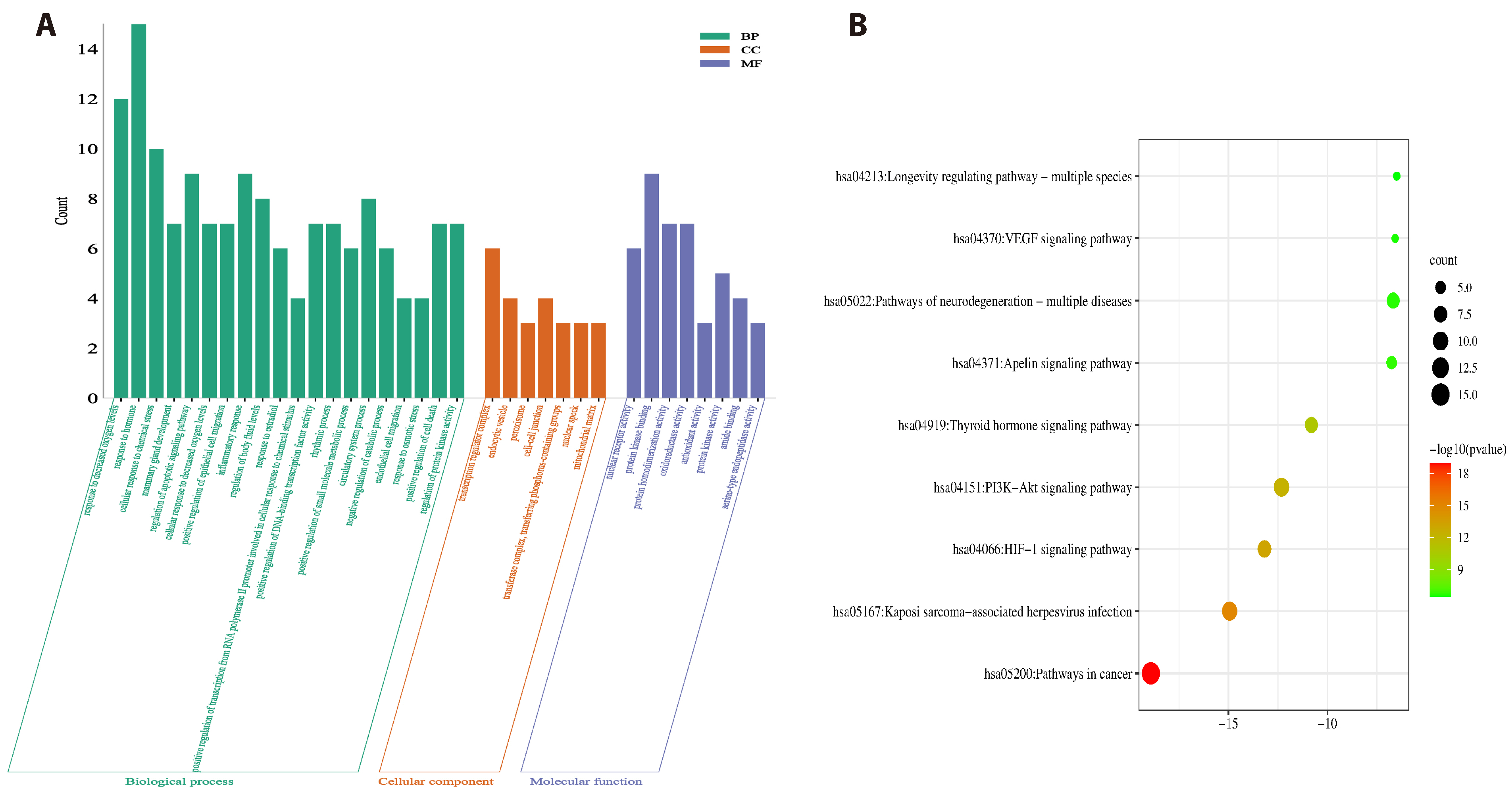
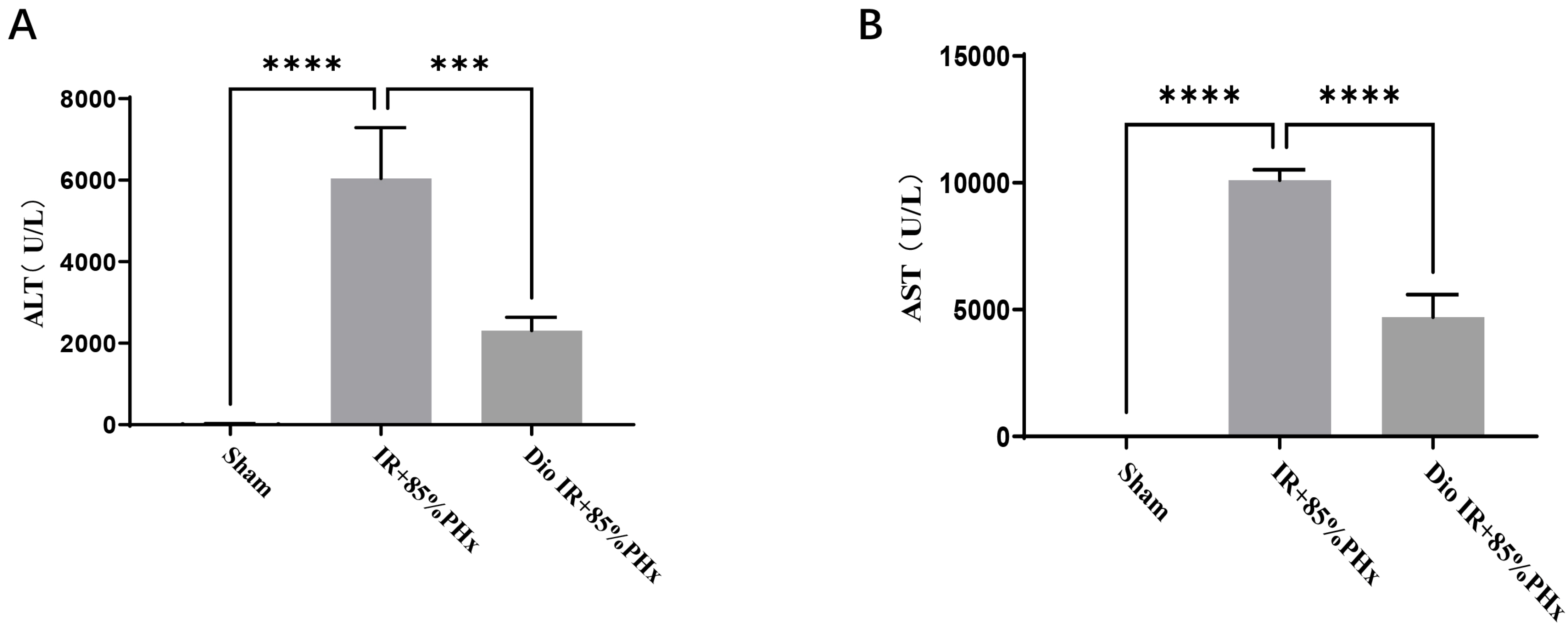
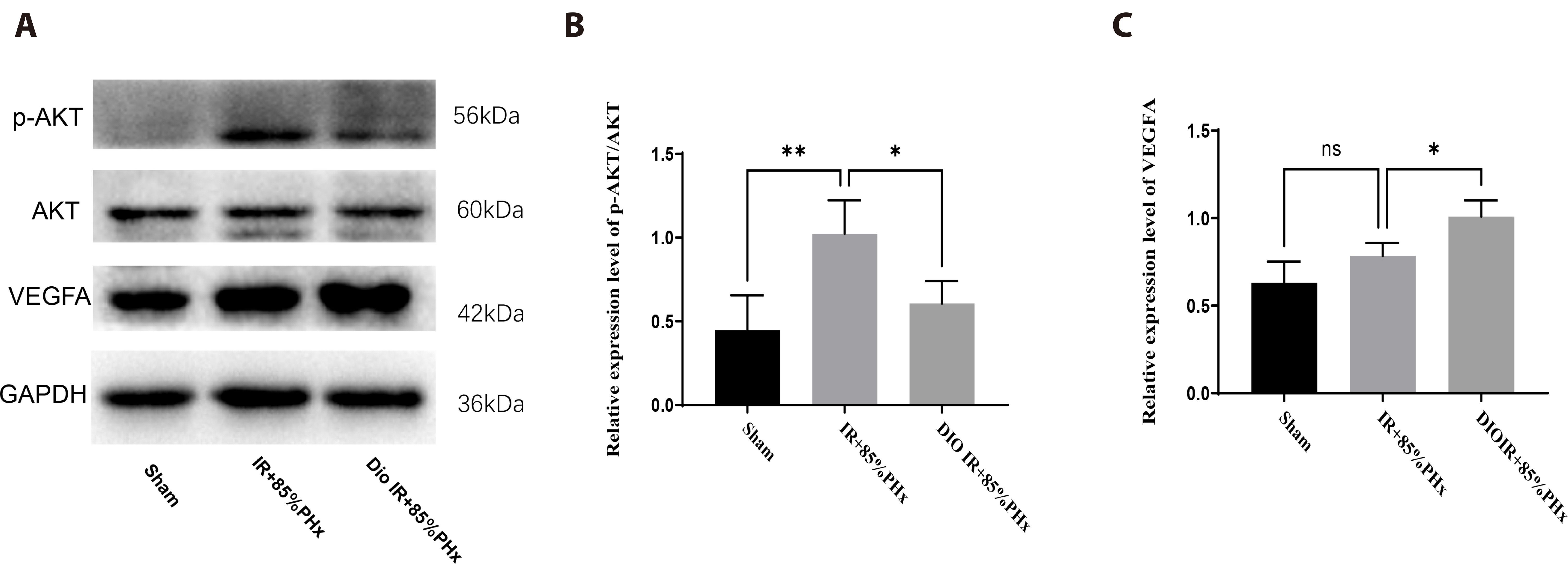
- Improving liver regeneration (LR) remains a medical issue, and there is currently a lack of safe and effective drugs for LR. Rhizoma Dioscoreae (SanYak, SY) is a traditional Chinese medicine. However, the underlying action mechanism of SY treatment for LR is yet to be fully elucidated. To explore the mechanism by which SY affects LR, we have conducted a series of methods for network pharmacological analysis, molecular docking, and in vivo experimental validation in mice. Overall, 9 compounds and 30 predicted target genes of SY were found to be associated with the therapeutic effects of LR. Compared with the model group, hematoxylin and eosin staining revealed that the mice with preoperative drug intervention possessed fewer postoperative hepatocyte bubbles and relatively regular morphology. Furthermore, the serum alanine transaminase and aspartate aminotransferase levels were reduced, immunohistochemistry revealed elevated proliferating cell nuclear antigen positivity rate, and Western blotting demonstrated that the phospho-protein kinase B (AKT)/AKT ratio was downregulated and that vascular endothelial growth factor A (VEGFA) expression levels were upregulated. This study explored dioscin, the main active ingredient of SY, and its potential therapeutic effects on LR. It repairs damaged liver following surgery and promotes liver cell proliferation. The action mechanism comprises reducing AKT phosphorylation levels and upregulating VEGFA expression levels. Thus, this study provides a new direction for further research on the mechanism of SY promoting LR.
Figure
Reference
-
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. 2021; Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 71:209–249. DOI: 10.3322/caac.21660. PMID: 33538338.2. Clavien PA, Oberkofler CE, Raptis DA, Lehmann K, Rickenbacher A, El-Badry AM. 2010; What is critical for liver surgery and partial liver transplantation: size or quality? Hepatology. 52:715–729. DOI: 10.1002/hep.23713. PMID: 20683967.3. Forbes SJ, Newsome PN. 2016; Liver regeneration - mechanisms and models to clinical application. Nat Rev Gastroenterol Hepatol. 13:473–485. DOI: 10.1038/nrgastro.2016.97. PMID: 27353402.4. Imai D, Maeda T, Wang H, Shimagaki T, Sanefuji K, Kayashima H, Tsutsui S, Matsuda H, Yoshizumi T, Mori M. 2021; Risk factors for and outcomes of intraoperative blood loss in liver resection for hepatocellular tumors. Am Surg. 87:376–383. DOI: 10.1177/0003134820949995. PMID: 32993315.5. Al-Saeedi M, Ghamarnejad O, Khajeh E, Shafiei S, Salehpour R, Golriz M, Mieth M, Weiss KH, Longerich T, Hoffmann K, Büchler MW, Mehrabi A. 2020; Pringle maneuver in extended liver resection: a propensity score analysis. Sci Rep. 10:8847. DOI: 10.1038/s41598-020-64596-y. PMID: 32483357. PMCID: PMC7264345.6. Teoh NC. 2011; Hepatic ischemia reperfusion injury: contemporary perspectives on pathogenic mechanisms and basis for hepatoprotection-the good, bad and deadly. J Gastroenterol Hepatol. 26 Suppl 1:180–187. DOI: 10.1111/j.1440-1746.2010.06584.x. PMID: 21199530.7. Martins PN, Theruvath TP, Neuhaus P. 2008; Rodent models of partial hepatectomies. Liver Int. 28:3–11. DOI: 10.1111/j.1478-3231.2007.01628.x. PMID: 18028319.8. Palmes D, Spiegel HU. 2004; Animal models of liver regeneration. Biomaterials. 25:1601–1611. DOI: 10.1016/S0142-9612(03)00508-8. PMID: 14697862.9. Mitchell C, Willenbring H. 2014; Addendum: a reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nat Protoc. 9:1532. DOI: 10.1038/nprot.2014.122. PMID: 24874817.10. Ando T, Hoshi M, Tezuka H, Ito H, Nakamoto K, Yamamoto Y, Saito K. 2023; Absence of indoleamine 2,3dioxygenase 2 promotes liver regeneration after partial hepatectomy in mice. Mol Med Rep. 27:24. DOI: 10.3892/mmr.2022.12911. PMID: 36484383. PMCID: PMC9813552.11. Zhou Q, Yu DH, Zhang N, Liu SM. 2019; Anti-inflammatory effect of total saponin fraction from Dioscorea nipponica Makino on gouty arthritis and its influence on NALP3 inflammasome. Chin J Integr Med. 25:663–670. DOI: 10.1007/s11655-016-2741-5. PMID: 28197935.12. Lee CY, Chou YE, Hsin MC, Lin CW, Wang PH, Yang SF, Hsiao YH. 2020; Dioscorea nipponica Makino suppresses TPA-induced migration and invasion through inhibition of matrix metalloproteinase-9 in human cervical cancer cells. Environ Toxicol. 35:1194–1201. DOI: 10.1002/tox.22984. PMID: 32519806.13. Liu C, Liao JZ, Li PY. 2017; Traditional Chinese herbal extracts inducing autophagy as a novel approach in therapy of nonalcoholic fatty liver disease. World J Gastroenterol. 23:1964–1973. DOI: 10.3748/wjg.v23.i11.1964. PMID: 28373762. PMCID: PMC5360637.14. Zuo H, Zhang Q, Su S, Chen Q, Yang F, Hu Y. 2018; A network pharmacology-based approach to analyse potential targets of traditional herbal formulas: an example of Yu Ping Feng decoction. Sci Rep. 8:11418. DOI: 10.1038/s41598-018-29764-1. PMID: 30061691. PMCID: PMC6065326.15. Zeng L, Yang K. 2017; Exploring the pharmacological mechanism of Yanghe Decoction on HER2-positive breast cancer by a network pharmacology approach. J Ethnopharmacol. 199:68–85. DOI: 10.1016/j.jep.2017.01.045. PMID: 28130113.16. Nacher JC, Schwartz JM. 2008; A global view of drug-therapy interactions. BMC Pharmacol. 8:5. DOI: 10.1186/1471-2210-8-5. PMID: 18318892. PMCID: PMC2294115.17. Ru J, Li P, Wang J, Zhou W, Li B, Huang C, Li P, Guo Z, Tao W, Yang Y, Xu X, Li Y, Wang Y, Yang L. 2014; TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 6:13. DOI: 10.1186/1758-2946-6-13. PMID: 24735618. PMCID: PMC4001360.18. UniProt Consortium. 2021; UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 49:D480–D489.19. Wang Y, Wang Q, Huang H, Huang W, Chen Y, McGarvey PB, Wu CH, Arighi CN. UniProt Consortium. 2021; A crowdsourcing open platform for literature curation in UniProt. PLoS Biol. 19:e3001464. DOI: 10.1371/journal.pbio.3001464. PMID: 34871295. PMCID: PMC8675915.20. Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, Stein TI, Nudel R, Lieder I, Mazor Y, Kaplan S, Dahary D, Warshawsky D, Guan-Golan Y, Kohn A, Rappaport N, Safran M, Lancet D. 2016; The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinformatics. 54:1. DOI: 10.1002/cpbi.5. PMID: 27322403.21. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003; Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13:2498–2504. DOI: 10.1101/gr.1239303. PMID: 14597658. PMCID: PMC403769.22. Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, Doncheva NT, Legeay M, Fang T, Bork P, Jensen LJ, von Mering C. 2021; Correction to 'The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets'. Nucleic Acids Res. 49:10800. Erratum for: Nucleic Acids Res. 2021;49:D605-D612. DOI: 10.1093/nar/gkaa1074. PMID: 33237311. PMCID: PMC7779004.23. Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, Chanda SK. 2019; Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 10:1523. DOI: 10.1038/s41467-019-09234-6. PMID: 30944313. PMCID: PMC6447622.24. Burley SK, Berman HM, Duarte JM, Feng Z, Flatt JW, Hudson BP, Lowe R, Peisach E, Piehl DW, Rose Y, Sali A, Sekharan M, Shao C, Vallat B, Voigt M, Westbrook JD, Young JY, Zardecki C. 2022; Protein Data Bank: a comprehensive review of 3D structure holdings and worldwide utilization by researchers, educators, and students. Biomolecules. 12:1425. DOI: 10.3390/biom12101425. PMID: 36291635. PMCID: PMC9599165.25. Burley SK, Bhikadiya C, Bi C, Bittrich S, Chen L, Crichlow GV, Christie CH, Dalenberg K, Di Costanzo L, Duarte JM, Dutta S, Feng Z, Ganesan S, Goodsell DS, Ghosh S, Green RK, Guranović V, Guzenko D, Hudson BP, Lawson CL, et al. 2021; RCSB Protein Data Bank: powerful new tools for exploring 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering and energy sciences. Nucleic Acids Res. 49:D437–D451. DOI: 10.1093/nar/gkaa1038. PMID: 33211854. PMCID: PMC7779003.26. DeLano WL. 2002; Unraveling hot spots in binding interfaces: progress and challenges. Curr Opin Struct Biol. 12:14–20. DOI: 10.1016/S0959-440X(02)00283-X. PMID: 11839484.27. Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. 2009; AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 30:2785–2791. DOI: 10.1002/jcc.21256. PMID: 19399780. PMCID: PMC2760638.28. Liu W, Shi Y, Cheng T, Jia R, Sun MZ, Liu S, Liu Q. 2021; Weighted gene coexpression network analysis in mouse livers following ischemia-reperfusion and extensive hepatectomy. Evid Based Complement Alternat Med. 2021:3897715. DOI: 10.1155/2021/3897715. PMID: 35003298. PMCID: PMC8736699.29. Wang S, Long S, Deng Z, Wu W. 2020; Positive role of Chinese herbal medicine in cancer immune regulation. Am J Chin Med. 48:1577–1592. DOI: 10.1142/S0192415X20500780. PMID: 33202152.30. Chen F, Zhong Z, Tan HY, Guo W, Zhang C, Tan CW, Li S, Wang N, Feng Y. 2020; Uncovering the anticancer mechanisms of Chinese herbal medicine formulas: therapeutic alternatives for liver cancer. Front Pharmacol. 11:293. DOI: 10.3389/fphar.2020.00293. PMID: 32256363. PMCID: PMC7093640.31. Yu S, Han B, Xing X, Li Y, Zhao D, Liu M, Wang S. 2021; A protein from Dioscorea polystachya (Chinese Yam) improves hydrocortisone-induced testicular dysfunction by alleviating leydig cell injury via upregulation of the Nrf2 pathway. Oxid Med Cell Longev. 2021:3575016. DOI: 10.1155/2021/3575016. PMID: 34887997. PMCID: PMC8651383.32. Meng X, Hu W, Wu S, Zhu Z, Lu R, Yang G, Qin C, Yang L, Nie G. 2019; Chinese yam peel enhances the immunity of the common carp (Cyprinus carpio L.) by improving the gut defence barrier and modulating the intestinal microflora. Fish Shellfish Immunol. 95:528–537. DOI: 10.1016/j.fsi.2019.10.066. PMID: 31678187.33. Zhang N, Liang T, Jin Q, Shen C, Zhang Y, Jing P. 2019; Chinese yam (Dioscorea opposita Thunb.) alleviates antibiotic-associated diarrhea, modifies intestinal microbiota, and increases the level of short-chain fatty acids in mice. Food Res Int. 122:191–198. DOI: 10.1016/j.foodres.2019.04.016. PMID: 31229072.34. Mohamadi-Zarch SM, Baluchnejadmojarad T, Nourabadi D, Khanizadeh AM, Roghani M. 2020; Protective effect of diosgenin on LPS/D-Gal-induced acute liver failure in C57BL/6 mice. Microb Pathog. 146:104243. DOI: 10.1016/j.micpath.2020.104243. PMID: 32389705.35. Feng X, Zhang Q, Li J, Bie N, Li C, Lian R, Qin L, Feng Y, Wang C. 2022; The impact of a novel Chinese yam-derived polysaccharide on blood glucose control in HFD and STZ-induced diabetic C57BL/6 mice. Food Funct. 13:2681–2692. DOI: 10.1039/D1FO03830C. PMID: 35170609.36. Wu MM, Wang QM, Huang BY, Mai CT, Wang CL, Wang TT, Zhang XJ. 2021; Dioscin ameliorates murine ulcerative colitis by regulating macrophage polarization. Pharmacol Res. 172:105796. DOI: 10.1016/j.phrs.2021.105796. PMID: 34343656.37. Liu P, Wang Z, Wei W. 2014; Phosphorylation of Akt at the C-terminal tail triggers Akt activation. Cell Cycle. 13:2162–2164. DOI: 10.4161/cc.29584. PMID: 24933731. PMCID: PMC4111671.38. Yang WL, Wu CY, Wu J, Lin HK. 2010; Regulation of Akt signaling activation by ubiquitination. Cell Cycle. 9:487–497. DOI: 10.4161/cc.9.3.10508. PMID: 20081374. PMCID: PMC3077544.39. Campana L, Esser H, Huch M, Forbes S. 2021; Liver regeneration and inflammation: from fundamental science to clinical applications. Nat Rev Mol Cell Biol. 22:608–624. DOI: 10.1038/s41580-021-00373-7. PMID: 34079104.40. Engelmann C, Habtesion A, Hassan M, Kerbert AJ, Hammerich L, Novelli S, Fidaleo M, Philips A, Davies N, Ferreira-Gonzalez S, Forbes SJ, Berg T, Andreola F, Jalan R. 2022; Combination of G-CSF and a TLR4 inhibitor reduce inflammation and promote regeneration in a mouse model of ACLF. J Hepatol. 77:1325–1338. DOI: 10.1016/j.jhep.2022.07.006. PMID: 35843375.41. Xiang M, Liu T, Tian C, Ma K, Gou J, Huang R, Li S, Li Q, Xu C, Li L, Lee CH, Zhang Y. 2022; Kinsenoside attenuates liver fibro-inflammation by suppressing dendritic cells via the PI3K-AKT-FoxO1 pathway. Pharmacol Res. 177:106092. DOI: 10.1016/j.phrs.2022.106092. PMID: 35066108. PMCID: PMC8776354.42. Zhong W, Qian K, Xiong J, Ma K, Wang A, Zou Y. 2016; Curcumin alleviates lipopolysaccharide induced sepsis and liver failure by suppression of oxidative stress-related inflammation via PI3K/AKT and NF-κB related signaling. Biomed Pharmacother. 83:302–313. DOI: 10.1016/j.biopha.2016.06.036. PMID: 27393927.43. Wu X, Luo Y, Wang S, Li Y, Bao M, Shang Y, Chen L, Liu W. 2022; AKAP12 ameliorates liver injury via targeting PI3K/AKT/PCSK6 pathway. Redox Biol. 53:102328. DOI: 10.1016/j.redox.2022.102328. PMID: 35576690. PMCID: PMC9118925.44. Amaravadi RK, Thompson CB. 2007; The roles of therapy-induced autophagy and necrosis in cancer treatment. Clin Cancer Res. 13:7271–7279. DOI: 10.1158/1078-0432.CCR-07-1595. PMID: 18094407.45. Yang J, Pi C, Wang G. 2018; Inhibition of PI3K/Akt/mTOR pathway by apigenin induces apoptosis and autophagy in hepatocellular carcinoma cells. Biomed Pharmacother. 103:699–707. DOI: 10.1016/j.biopha.2018.04.072. PMID: 29680738.46. Li H, Pang B, Nie B, Qu S, Zhang K, Xu J, Yang M, Liu J, Li S. 2022; Dioscin promotes autophagy by regulating the AMPK-mTOR pathway in ulcerative colitis. Immunopharmacol Immunotoxicol. 44:238–246. DOI: 10.1080/08923973.2022.2037632. PMID: 35174751.47. Yang Q, Wang C, Jin Y, Ma X, Xie T, Wang J, Liu K, Sun H. 2020; Corrigendum to "Disocin prevents postmenopausal atherosclerosis in ovariectomized LDLR-/mice through a PGC-1α/ERα pathway leading to promotion of autophagy and inhibition of oxidative stress, inflammation and apoptosis" [Pharmacol. Res. 148 (2019) 104414]. Pharmacol Res. 153:104523. Erratum for: Pharmacol Res. 2019;148:104414. DOI: 10.1016/j.phrs.2019.104523. PMID: 31926782.48. Bhardwaj N, Tripathi N, Goel B, Jain SK. 2021; Anticancer activity of diosgenin and its semi-synthetic derivatives: role in autophagy mediated cell death and induction of apoptosis. Mini Rev Med Chem. 21:1646–1665. DOI: 10.2174/1389557521666210105111224. PMID: 33402081.49. Lamanilao GG, Dogan M, Patel PS, Azim S, Patel DS, Bhattacharya SK, Eason JD, Kuscu C, Kuscu C, Bajwa A. 2023; Key hepatoprotective roles of mitochondria in liver regeneration. Am J Physiol Gastrointest Liver Physiol. 324:G207–G218. DOI: 10.1152/ajpgi.00220.2022. PMID: 36648139. PMCID: PMC9988520.50. Hou X, Huang M, Zeng X, Zhang Y, Sun A, Wu Q, Zhu L, Zhao H, Liao Y. 2021; The role of TRPC6 in renal ischemia/reperfusion and cellular hypoxia/reoxygenation injuries. Front Mol Biosci. 8:698975. DOI: 10.3389/fmolb.2021.698975. PMID: 34307458. PMCID: PMC8295989.51. Zhang X, Wang X, Khurm M, Zhan G, Zhang H, Ito Y, Guo Z. 2020; Alterations of brain quantitative proteomics profiling revealed the molecular mechanisms of diosgenin against cerebral ischemia reperfusion effects. J Proteome Res. 19:1154–1168. DOI: 10.1021/acs.jproteome.9b00667. PMID: 31940440.52. Zhang X, Wang X, Xue Z, Zhan G, Ito Y, Guo Z. 2021; Prevention properties on cerebral ischemia reperfusion of medicine food homologous Dioscorea yam-derived diosgenin based on mediation of potential targets. Food Chem. 345:128672. DOI: 10.1016/j.foodchem.2020.128672. PMID: 33352403.53. Zhang X, Xue Z, Zhu S, Guo Y, Zhang Y, Dou J, Zhang J, Ito Y, Guo Z. 2022; Diosgenin revealed potential effect against cerebral ischemia reperfusion through HIKESHI/HSP70/NF-κB anti-inflammatory axis. Phytomedicine. 99:153991. DOI: 10.1016/j.phymed.2022.153991. PMID: 35217435.54. 2020; Correction to: heart disease and stroke statistics-2019 update: a report from the American Heart Association. Circulation. 141:e33. Erratum for: Circulation. 2019;139:e56-e528. DOI: 10.1161/CIR.0000000000000746.55. Ding BS, Nolan DJ, Butler JM, James D, Babazadeh AO, Rosenwaks Z, Mittal V, Kobayashi H, Shido K, Lyden D, Sato TN, Rabbany SY, Rafii S. 2010; Inductive angiocrine signals from sinusoidal endothelium are required for liver regeneration. Nature. 468:310–315. DOI: 10.1038/nature09493. PMID: 21068842. PMCID: PMC3058628.56. Tian J, Cheng L, Kong E, Gu W, Jiang Y, Hao Q, Kong B, Sun L. 2022; linc00958/miR-185-5p/RSF-1 modulates cisplatin resistance and angiogenesis through AKT1/GSK3β/VEGFA pathway in cervical cancer. Reprod Biol Endocrinol. 20:132. DOI: 10.1186/s12958-022-00995-2. PMID: 36056431. PMCID: PMC9438131.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Mechanism of Wenshen Xuanbi Decoction in the treatment of osteoarthritis based on network pharmacology and experimental verification
- Leaf Spot of Yam Caused by Pseudophloeosporella dioscoreae in Korea
- EFFECTS OF EXTRACTS OF DRYNARIAE RHIZOMA ON THE CHARACTERISTICS OF RAT CALARIA AND BONE MARROW CELLS
- Adult Stem Cell Therapy in Chronic Liver Diseases
- Immunomodulatory Effects of Dioscoreae Rhizome Against Inflammation through Suppressed Production of Cytokines Via Inhibition of the NF-kappaB Pathway