Int J Stem Cells.
2024 May;17(2):194-203. 10.15283/ijsc23167.
Energy Metabolism in Human Pluripotent Stem and Differentiated Cells Compared Using a Seahorse XF96 Extracellular Flux Analyzer
- Affiliations
-
- 1Soonchunhyang Institute of Medi-bio Science (SIMS), Soon Chun Hyang University, Cheonan, Korea
- 2Dementia Research Group, Korea Brain Research Institute (KBRI), Daegu, Korea
- 3Department of Animal Science and Technology College of Biotechnology, Chung-Ang University, Anseong, Korea
- 4Department of Neurosurgery, Uijeonbu St. Mary’s Hospital, College of Medicine, The Catholic University of Korea, Seoul, Korea
- KMID: 2556237
- DOI: http://doi.org/10.15283/ijsc23167
Abstract
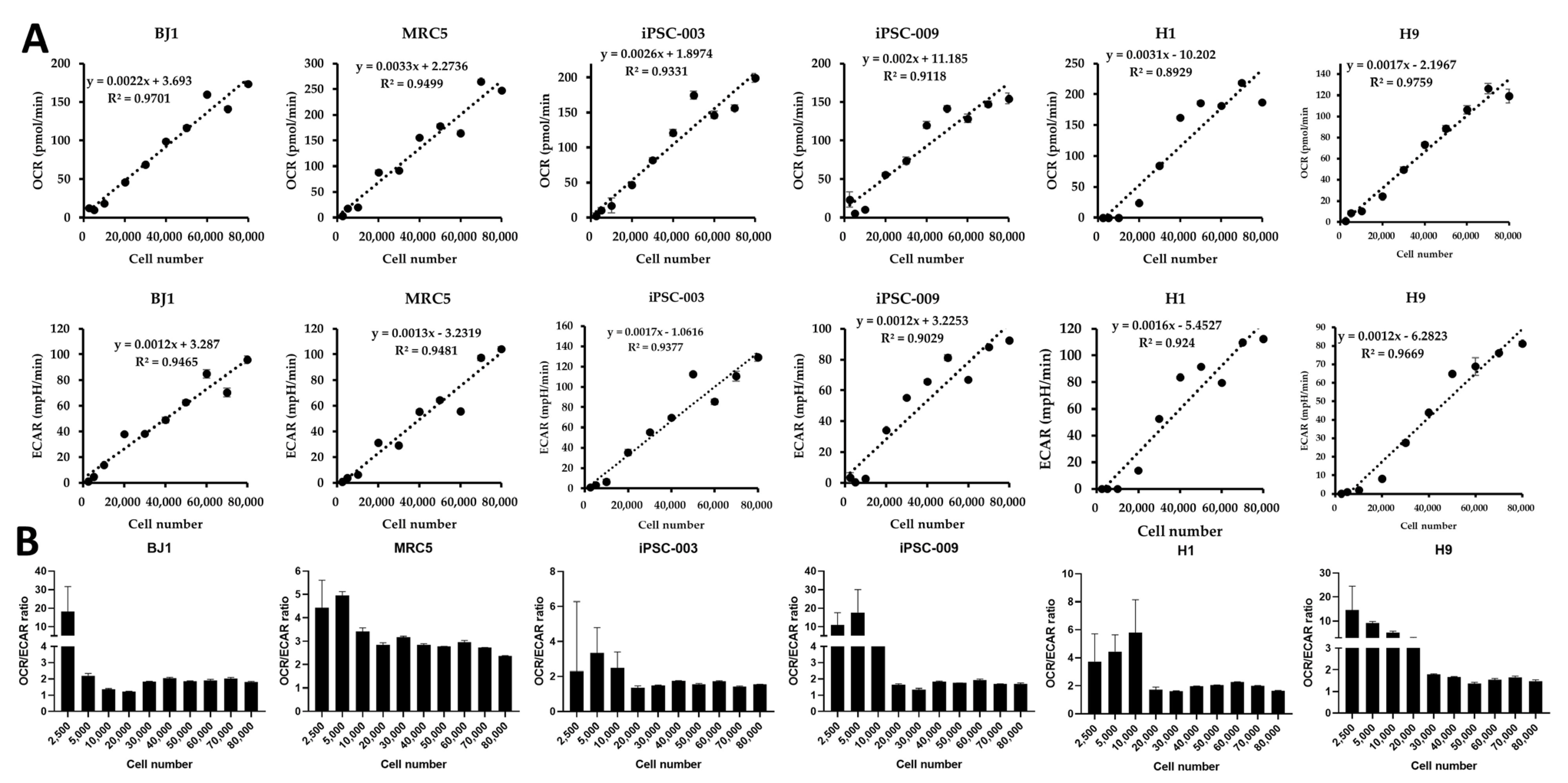
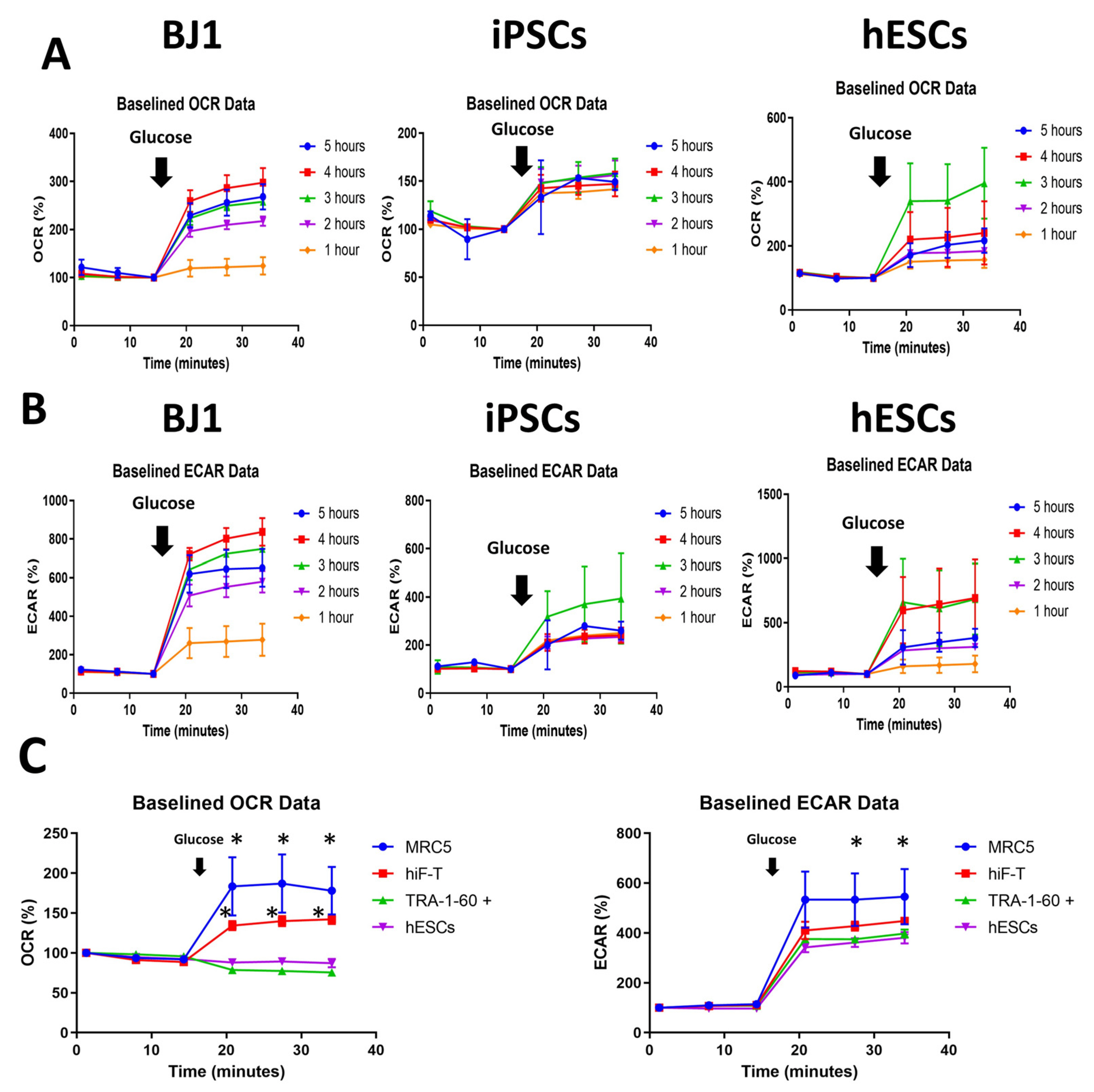
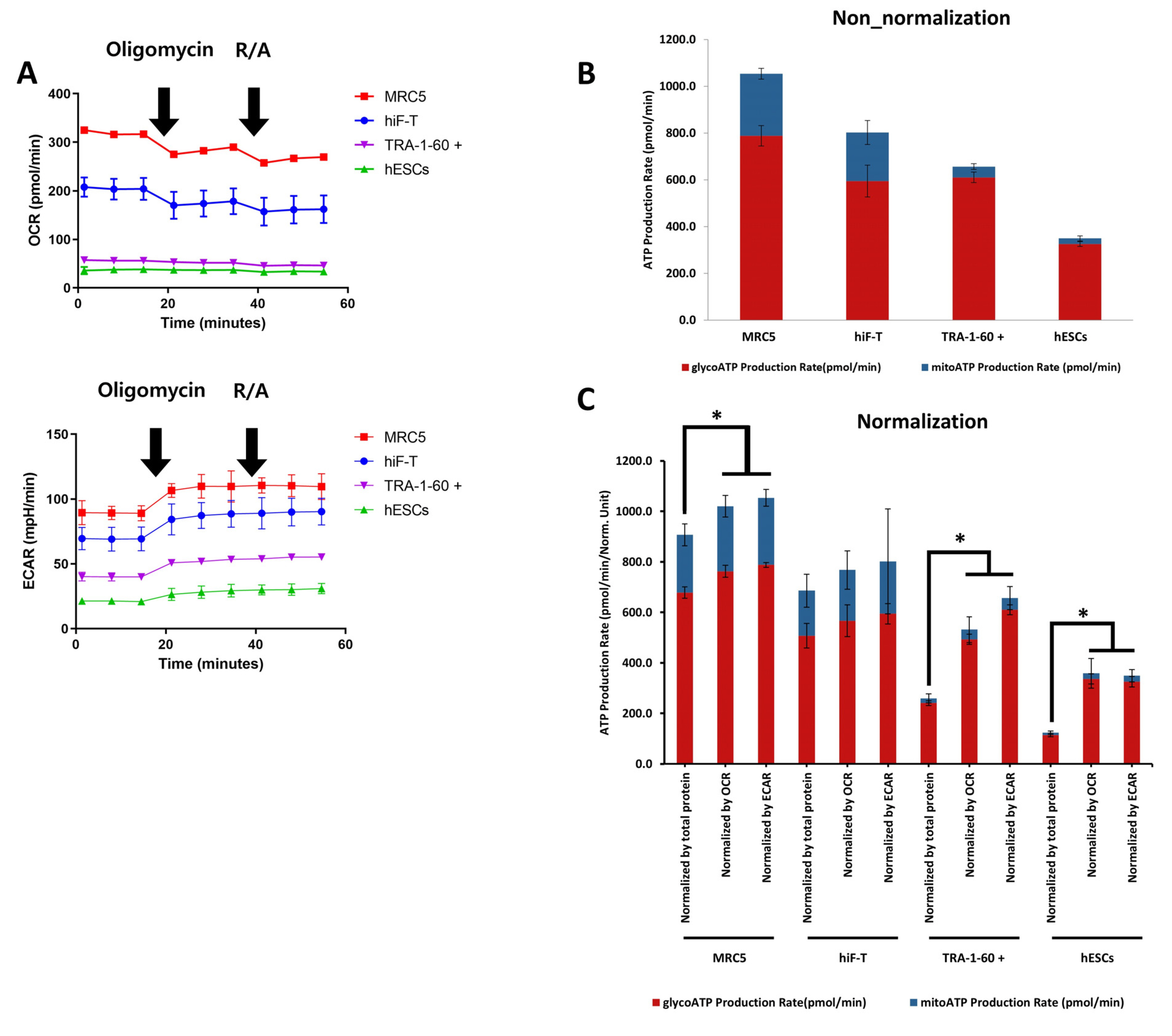
- Evaluating cell metabolism is crucial during pluripotent stem cell (PSC) differentiation and somatic cell reprogramming as it affects cell fate. As cultured stem cells are heterogeneous, a comparative analysis of relative metabolism using existing metabolic analysis methods is difficult, resulting in inaccuracies. In this study, we measured human PSC basal metabolic levels using a Seahorse analyzer. We used fibroblasts, human induced PSCs, and human embryonic stem cells to monitor changes in basal metabolic levels according to cell number and determine the number of cells suitable for analysis. We evaluated normalization methods using glucose and selected the most suitable for the metabolic analysis of heterogeneous PSCs during the reprogramming stage. The response of fibroblasts to glucose increased with starvation time, with oxygen consumption rate and extracellular acidification rate responding most effectively to glucose 4 hours after starvation and declining after 5 hours of starvation. Fibroblasts and PSCs achieved appropriate responses to glucose without damaging their metabolism 2∼4 and 2∼3 hours after starvation, respectively. We developed a novel method for comparing basal metabolic rates of fibroblasts and PSCs, focusing on quantitative analysis of glycolysis and oxidative phosphorylation using glucose without enzyme inhibitors. This protocol enables efficient comparison of energy metabolism among cell types, including undifferentiated PSCs, differentiated cells, and cells undergoing cellular reprogramming, and addresses critical issues, such as differences in basal metabolic levels and sensitivity to normalization, providing valuable insights into cellular energetics.
Figure
Reference
-
References
1. Zhang J, Nuebel E, Daley GQ, Koehler CM, Teitell MA. 2012; Metabolic regulation in pluripotent stem cells during reprogramming and self-renewal. Cell Stem Cell. 11:589–595. DOI: 10.1016/j.stem.2012.10.005. PMID: 23122286. PMCID: PMC3492890.
Article2. Nishimura K, Fukuda A, Hisatake K. 2019; Mechanisms of the metabolic shift during somatic cell reprogramming. Int J Mol Sci. 20:2254. DOI: 10.3390/ijms20092254. PMID: 31067778. PMCID: PMC6539623.
Article3. Agathocleous M, Love NK, Randlett O, et al. 2012; Metabolic differentiation in the embryonic retina. Nat Cell Biol. 14:859–864. DOI: 10.1038/ncb2531. PMID: 22750943. PMCID: PMC3442239.
Article4. Facucho-Oliveira JM, Alderson J, Spikings EC, Egginton S, St John JC. 2007; Mitochondrial DNA replication during differentiation of murine embryonic stem cells. J Cell Sci. 120(Pt 22):4025–4034. DOI: 10.1242/jcs.016972. PMID: 17971411.
Article5. Prigione A, Fauler B, Lurz R, Lehrach H, Adjaye J. 2010; The senescence-related mitochondrial/oxidative stress pathway is repressed in human induced pluripotent stem cells. Stem Cells. 28:721–733. DOI: 10.1002/stem.404. PMID: 20201066.
Article6. Suhr ST, Chang EA, Tjong J, et al. 2010; Mitochondrial rejuvenation after induced pluripotency. PLoS One. 5:e14095. DOI: 10.1371/journal.pone.0014095. PMID: 21124794. PMCID: PMC2991355.
Article7. Xu X, Duan S, Yi F, Ocampo A, Liu GH, Izpisua Belmonte JC. 2013; Mitochondrial regulation in pluripotent stem cells. Cell Metab. 18:325–332. DOI: 10.1016/j.cmet.2013.06.005. PMID: 23850316.
Article8. Carey BW, Finley LW, Cross JR, Allis CD, Thompson CB. 2015; Intracellular α-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature. 518:413–416. DOI: 10.1038/nature13981. PMID: 25487152. PMCID: PMC4336218.
Article9. Moussaieff A, Kogan NM, Aberdam D. 2015; Concise review: energy metabolites: key mediators of the epigenetic state of pluripotency. Stem Cells. 33:2374–2380. DOI: 10.1002/stem.2041. PMID: 25873344.
Article10. Shiraki N, Shiraki Y, Tsuyama T, et al. 2014; Methionine metabolism regulates maintenance and differentiation of human pluripotent stem cells. Cell Metab. 19:780–794. DOI: 10.1016/j.cmet.2014.03.017. PMID: 24746804.
Article11. Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. 2009; ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 324:1076–1080. DOI: 10.1126/science.1164097. PMID: 19461003. PMCID: PMC2746744.
Article12. Lunt SY, Vander Heiden MG. 2011; Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol. 27:441–464. DOI: 10.1146/annurev-cellbio-092910-154237. PMID: 21985671.
Article13. Ryall JG, Cliff T, Dalton S, Sartorelli V. 2015; Metabolic reprogramming of stem cell epigenetics. Cell Stem Cell. 17:651–662. DOI: 10.1016/j.stem.2015.11.012. PMID: 26637942. PMCID: PMC4672395.
Article14. Teslaa T, Teitell MA. 2015; Pluripotent stem cell energy metabolism: an update. EMBO J. 34:138–153. DOI: 10.15252/embj.201490446. PMID: 25476451. PMCID: PMC4337063.
Article15. Madonna R, Görbe A, Ferdinandy P, De Caterina R. 2013; Glucose metabolism, hyperosmotic stress, and reprogramming of somatic cells. Mol Biotechnol. 55:169–178. DOI: 10.1007/s12033-013-9668-2. PMID: 23657997.
Article16. Cha Y, Han MJ, Cha HJ, et al. 2017; Metabolic control of primed human pluripotent stem cell fate and function by the miR-200c-SIRT2 axis. Nat Cell Biol. 19:445–456. DOI: 10.1038/ncb3517. PMID: 28436968. PMCID: PMC5545746.
Article17. Kuo TC, Huang KY, Yang SC, et al. 2020; Monocarboxylate transporter 4 is a therapeutic target in non-small cell lung cancer with aerobic glycolysis preference. Mol Ther Oncolytics. 18:189–201. DOI: 10.1016/j.omto.2020.06.012. PMID: 32695876. PMCID: PMC7364124.
Article18. Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. 2002. Molecular biology of the cell. 4th ed. Garland Science.19. Tsogtbaatar E, Landin C, Minter-Dykhouse K, Folmes CDL. 2020; Energy metabolism regulates stem cell pluripotency. Front Cell Dev Biol. 8:87. DOI: 10.3389/fcell.2020.00087. PMID: 32181250. PMCID: PMC7059177.
Article20. Gu W, Gaeta X, Sahakyan A, et al. 2016; Glycolytic metabolism plays a functional role in regulating human pluripotent stem cell state. Cell Stem Cell. 19:476–490. DOI: 10.1016/j.stem.2016.08.008. PMID: 27618217. PMCID: PMC5055460.
Article21. Lees JG, Cliff TS, Gammilonghi A, et al. 2019; Oxygen regulates human pluripotent stem cell metabolic flux. Stem Cells Int. 2019:8195614. DOI: 10.1155/2019/8195614. PMID: 31236115. PMCID: PMC6545818.
Article22. Vander Heiden MG, Cantley LC, Thompson CB. 2009; Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 324:1029–1033. DOI: 10.1126/science.1160809. PMID: 19460998. PMCID: PMC2849637.
Article23. DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. 2008; The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 7:11–20. DOI: 10.1016/j.cmet.2007.10.002. PMID: 18177721.
Article24. Takahashi K, Tanabe K, Ohnuki M, et al. 2007; Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 131:861–872. DOI: 10.1016/j.cell.2007.11.019. PMID: 18035408.
Article25. Tachibana M, Amato P, Sparman M, et al. 2013; Human embryonic stem cells derived by somatic cell nuclear transfer. Cell. 153:1228–1238. DOI: 10.1016/j.cell.2013.05.006. PMID: 23683578. PMCID: PMC3772789.
Article26. Wang Q, Xiong Y, Zhang S, et al. 2021; The dynamics of metabolic characterization in iPSC-derived kidney organoid differentiation via a comparative omics approach. Front Genet. 12:632810. DOI: 10.3389/fgene.2021.632810. PMID: 33643392. PMCID: PMC7902935.
Article27. Kim YH, Heo JS, Han HJ. 2006; High glucose increase cell cycle regulatory proteins level of mouse embryonic stem cells via PI3-K/Akt and MAPKs signal pathways. J Cell Physiol. 209:94–102. DOI: 10.1002/jcp.20706. PMID: 16775839.
Article28. Folmes CD, Dzeja PP, Nelson TJ, Terzic A. 2012; Metabolic plasticity in stem cell homeostasis and differentiation. Cell Stem Cell. 11:596–606. DOI: 10.1016/j.stem.2012.10.002. PMID: 23122287. PMCID: PMC3593051.
Article29. Folmes CD, Nelson TJ, Martinez-Fernandez A, et al. 2011; Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metab. 14:264–271. DOI: 10.1016/j.cmet.2011.06.011. PMID: 21803296. PMCID: PMC3156138.
Article30. Kondoh H, Lleonart ME, Nakashima Y, et al. 2007; A high glycolytic flux supports the proliferative potential of murine embryonic stem cells. Antioxid Redox Signal. 9:293–299. DOI: 10.1089/ars.2006.1467. PMID: 17184172.
Article31. Prieto J, García-Cañaveras JC, León M, et al. 2021; c-MYC triggers lipid remodelling during early somatic cell reprogramming to pluripotency. Stem Cell Rev Rep. 17:2245–2261. DOI: 10.1007/s12015-021-10239-2. PMID: 34476741. PMCID: PMC8599373.
Article32. Gu X, Ma Y, Liu Y, Wan Q. 2020; Measurement of mitochondrial respiration in adherent cells by Seahorse XF96 Cell Mito Stress Test. STAR Protoc. 2:100245. DOI: 10.1016/j.xpro.2020.100245. PMID: 33458707. PMCID: PMC7797920.
Article33. Zhang J, Nuebel E, Wisidagama DR, et al. 2012; Measuring energy metabolism in cultured cells, including human pluripotent stem cells and differentiated cells. Nat Protoc. 7:1068–1085. DOI: 10.1038/nprot.2012.048. PMID: 22576106. PMCID: PMC3819135.
Article34. Ruas JS, Siqueira-Santos ES, Amigo I, Rodrigues-Silva E, Kowaltowski AJ, Castilho RF. 2016; Underestimation of the maximal capacity of the mitochondrial electron transport system in oligomycin-treated cells. PLoS One. 11:e0150967. DOI: 10.1371/journal.pone.0150967. PMID: 26950698. PMCID: PMC4780810.
Article35. Plitzko B, Loesgen S. 2018; Measurement of oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) in culture cells for assessment of the energy metabolism. Bio Protoc. 8:e2850. DOI: 10.21769/BioProtoc.2850. PMID: 34285967. PMCID: PMC8275291.
Article36. Schmidt CA, Fisher-Wellman KH, Neufer PD. 2021; From OCR and ECAR to energy: perspectives on the design and interpretation of bioenergetics studies. J Biol Chem. 297:101140. DOI: 10.1016/j.jbc.2021.101140. PMID: 34461088. PMCID: PMC8479256.
Article37. Kim HK, Ha TW, Lee MR. 2021; Single-cell transcriptome analysis as a promising tool to study pluripotent stem cell reprogramming. Int J Mol Sci. 22:5988. DOI: 10.3390/ijms22115988. PMID: 34206025. PMCID: PMC8198005.
Article38. Zhang C, Skamagki M, Liu Z, et al. 2017; Biological significance of the suppression of oxidative phosphorylation in induced pluripotent stem cells. Cell Rep. 21:2058–2065. DOI: 10.1016/j.celrep.2017.10.098. PMID: 29166598. PMCID: PMC5841608.
Article39. Yu L, Ji KY, Zhang J, et al. 2019; Core pluripotency factors promote glycolysis of human embryonic stem cells by activating GLUT1 enhancer. Protein Cell. 10:668–680. DOI: 10.1007/s13238-019-0637-9. PMID: 31152430. PMCID: PMC6711954.
Article40. Cacchiarelli D, Trapnell C, Ziller MJ, et al. 2015; Integrative analyses of human reprogramming reveal dynamic nature of induced pluripotency. Cell. 162:412–424. DOI: 10.1016/j.cell.2015.06.016. PMID: 26186193. PMCID: PMC4511597.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Engineering Biomaterials for Feeder-Free Maintenance of Human Pluripotent Stem Cells
- Stem Cell Properties of Therapeutic Potential
- Neuronal Differentiation of a Human Induced Pluripotent Stem Cell Line (FS-1) Derived from Newborn Foreskin Fibroblasts
- Human Placenta-Derived ECM Supports Tri-Lineage Differentiation of Human Induced Pluripotent Stem Cells
- Disease Specific Induced Pluripotent Stem Cells by Using Transcription Factors