J Stroke.
2023 Sep;25(3):350-360. 10.5853/jos.2023.00381.
Dissecting Causal Relationships Between Gut Microbiota, Blood Metabolites, and Stroke: A Mendelian Randomization Study
- Affiliations
-
- 1Department of Endocrine and Metabolic Diseases, Shanghai Institute of Endocrine and Metabolic Diseases, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China
- 2Shanghai National Clinical Research Center for Metabolic Diseases, Key Laboratory for Endocrine and Metabolic Diseases of the National Health Commission of the PR China, Shanghai Key Laboratory for Endocrine Tumor, State Key Laboratory of Medical Genomics, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China
- 3MRC Integrative Epidemiology Unit, Bristol Medical School, University of Bristol, Bristol, UK
- KMID: 2546436
- DOI: http://doi.org/10.5853/jos.2023.00381
Abstract
- Background and Purpose
We investigated the causal relationships between the gut microbiota (GM), stroke, and potential metabolite mediators using Mendelian randomization (MR).
Methods
We leveraged the summary statistics of GM (n=18,340 in the MiBioGen consortium), blood metabolites (n=115,078 in the UK Biobank), and stroke (cases n=60,176 and controls n=1,310,725 in the Global Biobank Meta-Analysis Initiative) from the largest genome-wide association studies to date. We performed bidirectional MR analyses to explore the causal relationships between the GM and stroke, and two mediation analyses, two-step MR and multivariable MR, to discover potential mediating metabolites.
Results
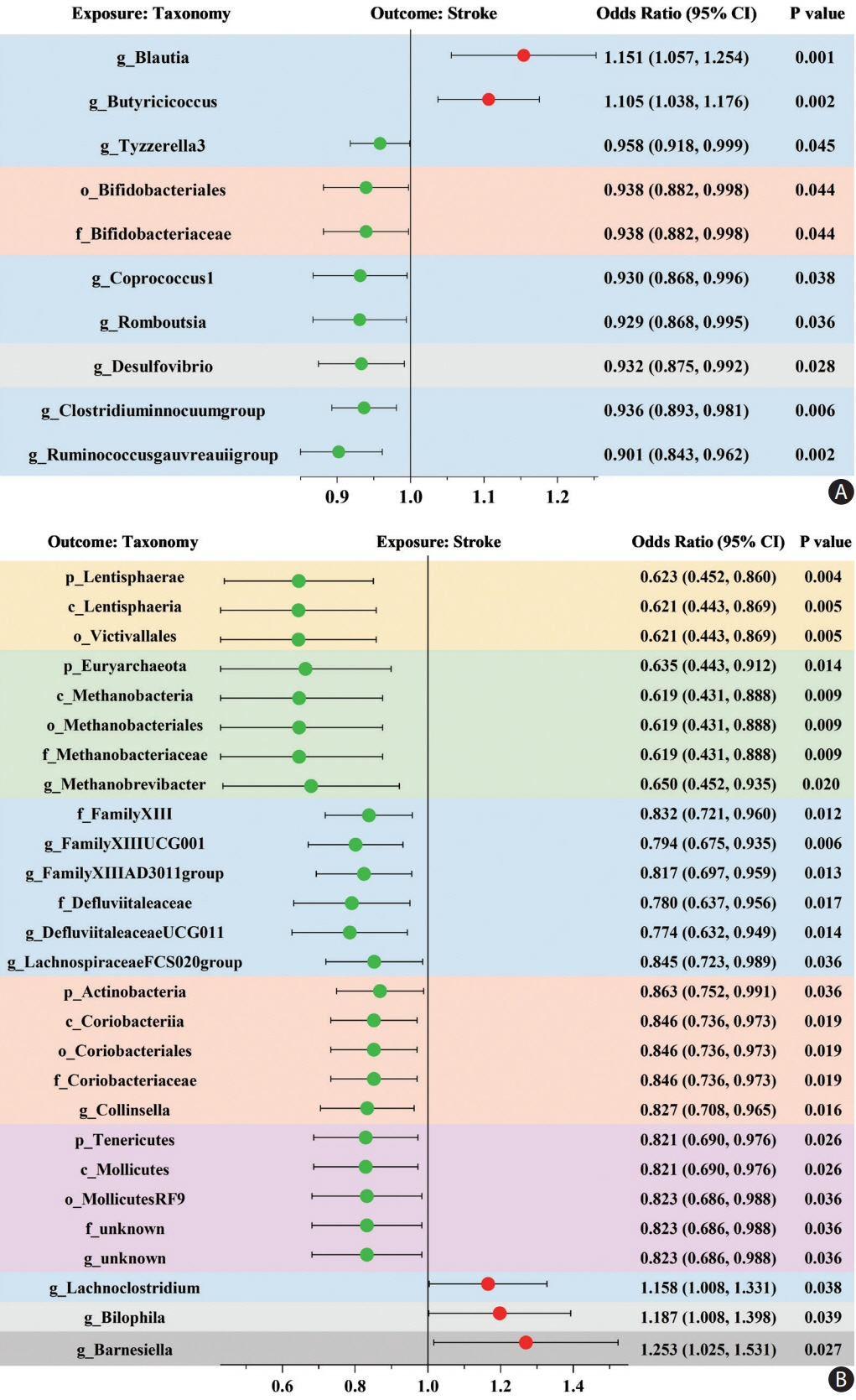
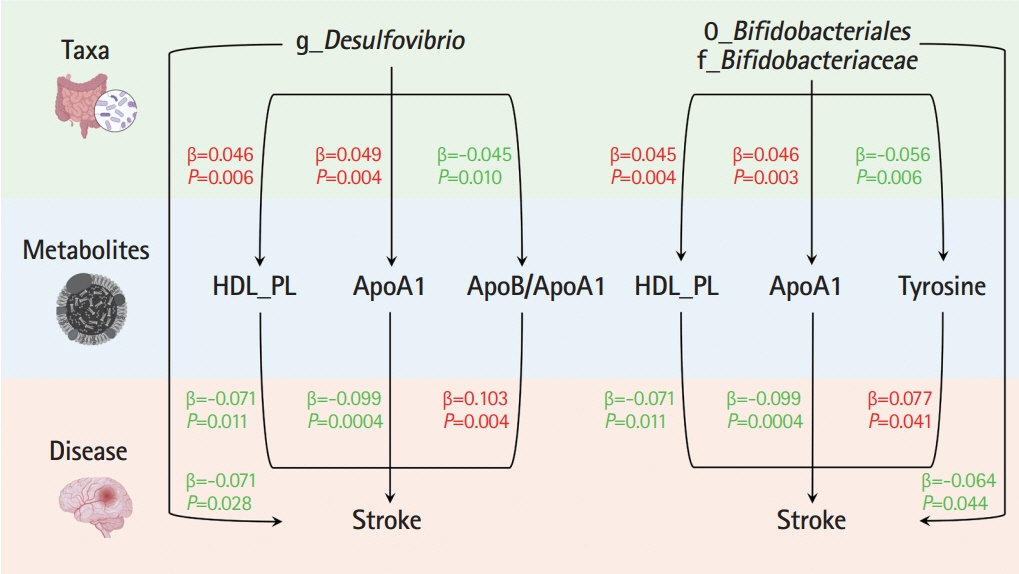
Ten taxa were causally associated with stroke, and stroke led to changes in 27 taxa. In the two-step MR, Bifidobacteriales order, Bifidobacteriaceae family, Desulfovibrio genus, apolipoprotein A1 (ApoA1), phospholipids in high-density lipoprotein (HDL_PL), and the ratio of apolipoprotein B to ApoA1 (ApoB/ApoA1) were causally associated with stroke (all P<0.044). The causal associations between Bifidobacteriales order, Bifidobacteriaceae family and stroke were validated using the weighted median method in an independent cohort. The three GM taxa were all positively associated with ApoA1 and HDL_PL, whereas Desulfovibrio genus was negatively associated with ApoB/ApoA1 (all P<0.010). Additionally, the causal associations between the three GM taxa and ApoA1 remained significant after correcting for the false discovery rate (all q-values <0.027). Multivariable MR showed that the associations between Bifidobacteriales order, Bifidobacteriaceae family and stroke were mediated by ApoA1 and HDL_PL, each accounting for 6.5% (P=0.028) and 4.6% (P=0.033); the association between Desulfovibrio genus and stroke was mediated by ApoA1, HDL_PL, and ApoB/ApoA1, with mediated proportions of 7.6% (P=0.019), 4.2% (P=0.035), and 9.1% (P=0.013), respectively.
Conclusion
The current MR study provides evidence supporting the causal relationships between several specific GM taxa and stroke and potential mediating metabolites.
Figure
Reference
-
References
1. Tsao CW, Aday AW, Almarzooq ZI, Alonso A, Beaton AZ, Bittencourt MS, et al. Heart disease and stroke statistics-2022 update: a report from the American Heart Association. Circulation. 2022; 145:e153–e639.2. Feigin VL, Norrving B, Mensah GA. Global burden of stroke. Circ Res. 2017; 120:439–448.3. Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, et al. Host-gut microbiota metabolic interactions. Science. 2012; 336:1262–1267.4. Honarpisheh P, Bryan RM, McCullough LD. Aging microbiotagut-brain axis in stroke risk and outcome. Circ Res. 2022; 130:1112–1144.5. Xu K, Gao X, Xia G, Chen M, Zeng N, Wang S, et al. Rapid gut dysbiosis induced by stroke exacerbates brain infarction in turn. Gut. 2021; 70:1486–1494.6. Winek K, Dirnagl U, Meisel A. Role of the gut microbiota in ischemic stroke. Neurol Int Open. 2017; 1:E287–E293.7. Yin J, Liao SX, He Y, Wang S, Xia GH, Liu FT, et al. Dysbiosis of gut microbiota with reduced trimethylamine-N-oxide level in patients with large-artery atherosclerotic stroke or transient ischemic attack. J Am Heart Assoc. 2015; 4:e002699.8. Xia GH, You C, Gao XX, Zeng XL, Zhu JJ, Xu KY, et al. Stroke dysbiosis index (SDI) in gut microbiome are associated with brain injury and prognosis of stroke. Front Neurol. 2019; 10:397.9. Chen Y, Liang J, Ouyang F, Chen X, Lu T, Jiang Z, et al. Persistence of gut microbiota dysbiosis and chronic systemic inflammation after cerebral infarction in cynomolgus monkeys. Front Neurol. 2019; 10:661.10. Singh V, Roth S, Llovera G, Sadler R, Garzetti D, Stecher B, et al. Microbiota dysbiosis controls the neuroinflammatory response after stroke. J Neurosci. 2016; 36:7428–7440.11. Peh A, O’Donnell JA, Broughton BRS, Marques FZ. Gut microbiota and their metabolites in stroke: a double-edged sword. Stroke. 2022; 53:1788–1801.12. Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med. 2008; 27:1133–1163.13. Sanna S, van Zuydam NR, Mahajan A, Kurilshikov A, Vich Vila A, Võsa U, et al. Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat Genet. 2019; 51:600–605.14. Skrivankova VW, Richmond RC, Woolf BAR, Yarmolinsky J, Davies NM, Swanson SA, et al. Strengthening the reporting of observational studies in epidemiology using Mendelian randomization: the STROBE-MR statement. JAMA. 2021; 326:1614–1621.15. Kurilshikov A, Medina-Gomez C, Bacigalupe R, Radjabzadeh D, Wang J, Demirkan A, et al. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat Genet. 2021; 53:156–165.16. Zhou W, Kanai M, Wu KH, Rasheed H, Tsuo K, Hirbo JB, et al. Global Biobank Meta-analysis Initiative: powering genetic discovery across human disease. Cell Genom. 2022; 2:100192.17. Julkunen H, Cichon´ska A, Tiainen M, Koskela H, Nybo K, Mäkelä V, et al. Atlas of plasma NMR biomarkers for health and disease in 118,461 individuals from the UK Biobank. Nat Commun. 2023; 14:604.18. Liu X, Tong X, Zou Y, Lin X, Zhao H, Tian L, et al. Mendelian randomization analyses support causal relationships between blood metabolites and the gut microbiome. Nat Genet. 2022; 54:52–61.19. Sudmant PH, Rausch T, Gardner EJ, Handsaker RE, Abyzov A, Huddleston J, et al. An integrated map of structural variation in 2,504 human genomes. Nature. 2015; 526:75–81.20. Burgess S, Thompson SG; CRP CHD Genetics Collaboration. Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol. 2011; 40:755–764.21. Burgess S, Small DS, Thompson SG. A review of instrumental variable estimators for Mendelian randomization. Stat Methods Med Res. 2017; 26:2333–2355.22. Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Patterson N, et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015; 47:291–295.23. Jia J, Dou P, Gao M, Kong X, Li C, Liu Z, et al. Assessment of causal direction between gut microbiota-dependent metabolites and cardiometabolic health: a bidirectional Mendelian randomization analysis. Diabetes. 2019; 68:1747–1755.24. Fu J, Bonder MJ, Cenit MC, Tigchelaar EF, Maatman A, Dekens JA, et al. The gut microbiome contributes to a substantial proportion of the variation in blood lipids. Circ Res. 2015; 117:817–824.25. Hindy G, Engström G, Larsson SC, Traylor M, Markus HS, Melander O, et al. Role of blood lipids in the development of ischemic stroke and its subtypes: a Mendelian randomization study. Stroke. 2018; 49:820–827.26. Burgess S, Daniel RM, Butterworth AS, Thompson SG; EPIC-InterAct Consortium. Network Mendelian randomization: using genetic variants as instrumental variables to investigate mediation in causal pathways. Int J Epidemiol. 2015; 44:484–495.27. Sanderson E. Multivariable Mendelian randomization and mediation. Cold Spring Harb Perspect Med. 2021; 11:a038984.28. Burgess S, Thompson SG. Multivariable Mendelian randomization: the use of pleiotropic genetic variants to estimate causal effects. Am J Epidemiol. 2015; 181:251–260.29. Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015; 44:512–525.30. Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018; 7:e34408.31. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B (Methodol). 1995; 57:289–300.32. Xu N, Kan P, Yao X, Yang P, Wang J, Xiang L, et al. Astragaloside IV reversed the autophagy and oxidative stress induced by the intestinal microbiota of AIS in mice. J Microbiol. 2018; 56:838–846.33. Chen YR, Jing QL, Chen FL, Zheng H, Chen LD, Yang ZC. Desulfovibrio is not always associated with adverse health effects in the Guangdong Gut Microbiome Project. PeerJ. 2021; 9:e12033.34. Mitsuoka T. Bifidobacteria and their role in human health. J Ind Microbiol. 1990; 6:263–267.35. Koskinen TT, Virtanen HEK, Voutilainen S, Tuomainen TP, Mursu J, Virtanen JK. Intake of fermented and non-fermented dairy products and risk of incident CHD: the Kuopio Ischaemic Heart Disease Risk Factor Study. Br J Nutr. 2018; 120:1288–1297.36. Li H, Zhang X, Pan D, Liu Y, Yan X, Tang Y, et al. Dysbiosis characteristics of gut microbiota in cerebral infarction patients. Transl Neurosci. 2020; 11:124–133.37. Liu Y, Kong C, Gong L, Zhang X, Zhu Y, Wang H, et al. The association of post-stroke cognitive impairment and gut microbiota and its corresponding metabolites. J Alzheimers Dis. 2020; 73:1455–1466.38. Dang Y, Zhang X, Zheng Y, Yu B, Pan D, Jiang X, et al. Distinctive gut microbiota alteration is associated with poststroke functional recovery: results from a prospective cohort study. Neural Plast. 2021; 2021:1469339.39. O’Donnell MJ, Chin SL, Rangarajan S, Xavier D, Liu L, Zhang H, et al. Global and regional effects of potentially modifiable risk factors associated with acute stroke in 32 countries (INTERSTROKE): a case-control study. Lancet. 2016; 388:761–775.40. Meikle PJ, Formosa MF, Mellett NA, Jayawardana KS, Giles C, Bertovic DA, et al. HDL phospholipids, but not cholesterol distinguish acute coronary syndrome from stable coronary artery disease. J Am Heart Assoc. 2019; 8:e011792.41. Yun KE, Kim J, Kim MH, Park E, Kim HL, Chang Y, et al. Major lipids, apolipoproteins, and alterations of gut microbiota. J Clin Med. 2020; 9:1589.42. Yiu JHC, Chan KS, Cheung J, Li J, Liu Y, Wang Y, et al. Gut microbiota-associated activation of TLR5 induces apolipoprotein A1 production in the liver. Circ Res. 2020; 127:1236–1252.43. Rühlemann MC, Hermes BM, Bang C, Doms S, Moitinho-Silva L, Thingholm LB, et al. Genome-wide association study in 8,956 German individuals identifies influence of ABO histo-blood groups on gut microbiome. Nat Genet. 2021; 53:147–155.44. Faust K, Raes J. Microbial interactions: from networks to models. Nat Rev Microbiol. 2012; 10:538–550.45. Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet. 2014; 23(R1):R89–R98.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- The Gut Microbiome and Osteoarthritis: A Two-Sample Mendelian Randomization Study
- Impaired pulmonary function mediates the impact of preterm birth on later-life stroke: a 2-step, multivariable Mendelian randomization study
- Clinical and genetic relationships between the QTc interval and risk of a stroke among atrial fibrillation patients undergoing catheter ablation
- The mediating role of atrial fibrillation in causal associations between risk factors and stroke: a Mendelian randomization study
- The Association between the Gut Microbiota and Erectile Dysfunction