J Rheum Dis.
2021 Apr;28(2):94-100. 10.4078/jrd.2021.28.2.94.
The Gut Microbiome and Osteoarthritis: A Two-Sample Mendelian Randomization Study
- Affiliations
-
- 1Department of Rheumatology, Korea University College of Medicine, Seoul, Korea
- KMID: 2514120
- DOI: http://doi.org/10.4078/jrd.2021.28.2.94
Abstract
Objective
The aim of this study was to examine if the intestinal microbiome is causally correlated with osteoarthritis (OA) incidence.
Methods
A two-sample Mendelian randomization (MR) study was conducted using inverse variance weighting (IVW), weighted median, and MR-Egger regression techniques. Publicly accessible summary statistics dataset of intestinal microbiomes of European descent from genome-wide association studies (GWASs) (a total with 3,326 individuals) was used as an exposure. As an outcome, summary data from the GWAS include 3,498 patients with OA of the knee and hip from the arcOGEN sample and 11,009 controls of European descent.
Results
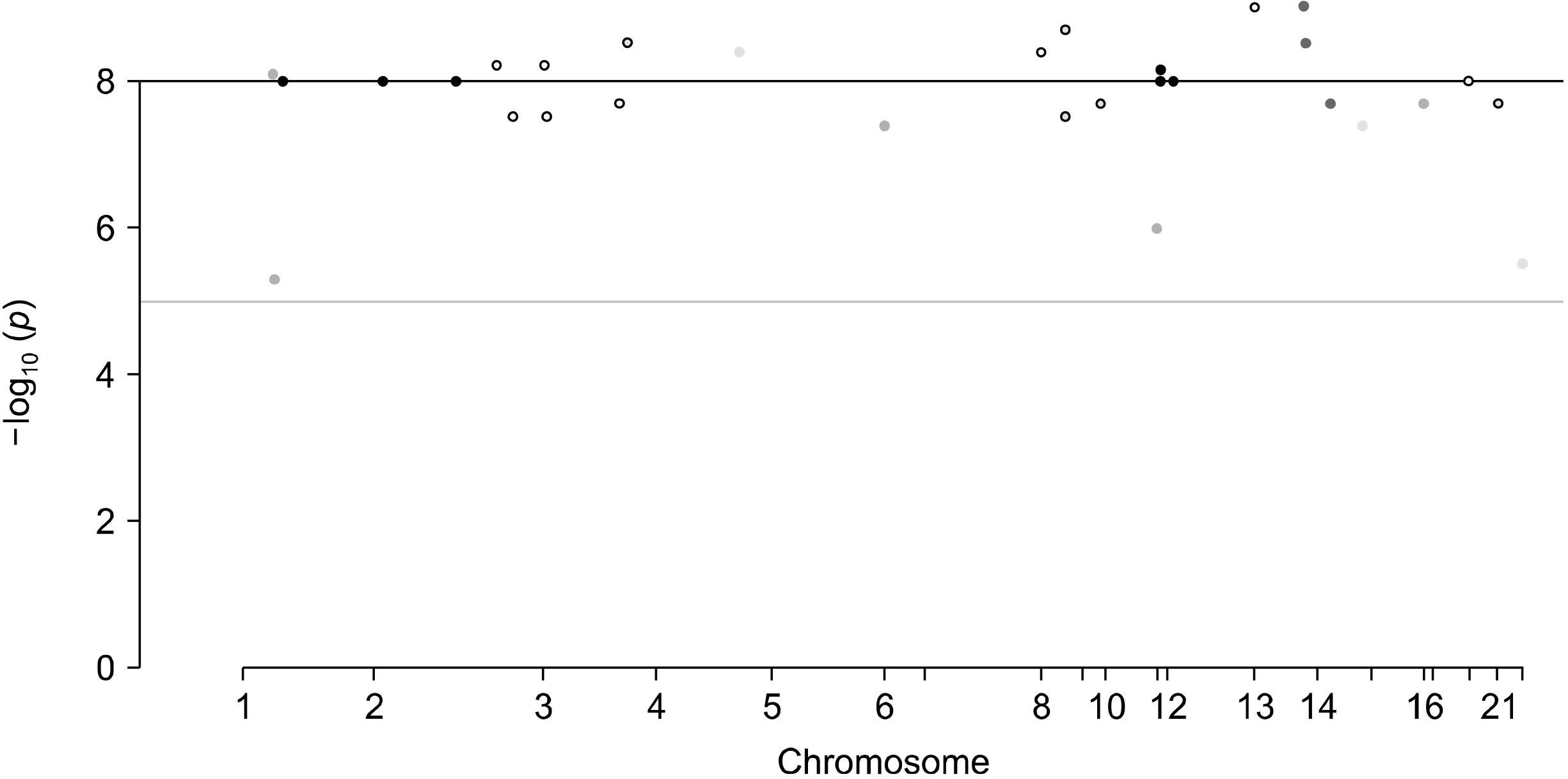
We identified 29 single-nucleotide polymorphisms from GWAS of intestinal microbiomes as instrumental variables. The IVW approach found no evidence to suggest a causal relationship between the intestinal microbiota and OA (beta=−0.001, standard error [SE]=0.004, p=0.748). The regression test of MR-Egger showed that the directional pleiotropy was unlikely to be a bias (intercept=0.002, SE=0.007, p=0.697) and the MR-Egger study showed no causal relation between the intestinal microbiota and the OA (beta=−0.002, SE=0.005, p=0.630). The weighted median analysis also did not have indications of a causal relationship between the intestinal microbiota and OA (beta=−0.002, SE=0.005, p=0.630). The MR results calculated using IVW, the median weighted and the MR-Egger regression approaches were consistent.
Conclusion
The findings of the MR analysis did not support a causal relationship between intestinal microbiome and OA risk.
Figure
Cited by 1 articles
-
Gut Microbiome Bridges Over Troubled Joints
Ji Won Kim, Ji Hyeon Ju
J Rheum Dis. 2021;28(3):111-112. doi: 10.4078/jrd.2021.28.3.111.
Reference
-
1. Dieppe PA, Lohmander LS. 2005; Pathogenesis and management of pain in osteoarthritis. Lancet. 365:965–73. DOI: 10.1016/S0140-6736(05)71086-2. PMID: 15766999.
Article2. Litwic A, Edwards MH, Dennison EM, Cooper C. 2013; Epidemiology and burden of osteoarthritis. Br Med Bull. 105:185–99. DOI: 10.1093/bmb/lds038. PMID: 23337796. PMCID: PMC3690438.
Article3. Berthelot JM, Sellam J, Maugars Y, Berenbaum F. 2019; Cartilage- gut-microbiome axis: a new paradigm for novel therapeutic opportunities in osteoarthritis. RMD Open. 5:e001037. DOI: 10.1136/rmdopen-2019-001037. PMID: 31673418. PMCID: PMC6803002.4. Favazzo LJ, Hendesi H, Villani DA, Soniwala S, Dar QA, Schott EM, et al. 2020; The gut microbiome-joint connection: implications in osteoarthritis. Curr Opin Rheumatol. 32:92–101. DOI: 10.1097/BOR.0000000000000681. PMID: 31724973. PMCID: PMC6903327.
Article5. Zhao Y, Chen B, Li S, Yang L, Zhu D, Wang Y, et al. 2018; Detection and characterization of bacterial nucleic acids in culture-negative synovial tissue and fluid samples from rheumatoid arthritis or osteoarthritis patients. Sci Rep. 8:14305. DOI: 10.1038/s41598-018-32675-w. PMID: 30250232. PMCID: PMC6155189.
Article6. Griffin TM, Huebner JL, Kraus VB, Yan Z, Guilak F. 2012; Induction of osteoarthritis and metabolic inflammation by a very high-fat diet in mice: effects of short-term exercise. Arthritis Rheum. 64:443–53. DOI: 10.1002/art.33332. PMID: 21953366. PMCID: PMC3268860.
Article7. Lorenzo D, GianVincenzo Z, Carlo Luca R, Karan G, Jorge V, Roberto M, et al. 2019; Oral-gut microbiota and arthritis: is there an evidence-based axis? J Clin Med. 8:1753. DOI: 10.3390/jcm8101753. PMID: 31652577. PMCID: PMC6832398.8. Ranstam J. 2008; Bias in observational studies. Acta Radiol. 49:644–5. DOI: 10.1080/02841850802075082. PMID: 18568556.
Article9. Burgess S, Daniel RM, Butterworth AS, Thompson SG. 2015; Network Mendelian randomization: using genetic variants as instrumental variables to investigate mediation in causal pathways. Int J Epidemiol. 44:484–95. DOI: 10.1093/ije/dyu176. PMID: 25150977. PMCID: PMC4469795.
Article10. Song GG, Lee YH. 2019; Causal association between bone mineral density and osteoarthritis: a Mendelian randomization study. J Rheum Dis. 26:104–10. DOI: 10.4078/jrd.2019.26.2.104.
Article11. Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, et al. 2014; The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 42(Database issue):D1001–6. DOI: 10.1093/nar/gkt1229. PMID: 24316577. PMCID: PMC3965119.
Article12. Wang J, Thingholm LB, Skiecevičienė J, Rausch P, Kummen M, Hov JR, et al. 2016; Genome-wide association analysis identifies variation in vitamin D receptor and other host factors influencing the gut microbiota. Nat Genet. 48:1396–406. DOI: 10.1038/ng.3695. PMID: 27723756. PMCID: PMC5626933.
Article13. Bonder MJ, Kurilshikov A, Tigchelaar EF, Mujagic Z, Imhann F, Vila AV, et al. 2016; The effect of host genetics on the gut microbiome. Nat Genet. 48:1407–12. DOI: 10.1038/ng.3663. PMID: 27694959.
Article14. Zeggini E, Panoutsopoulou K, Southam L, Rayner NW, Day-Williams AG, Lopes MC, et al. arcOGEN Consortium and arcOGEN Collaborators. 2012; Identification of new susceptibility loci for osteoarthritis (arcOGEN): a genome-wide association study. Lancet. 380:815–23. DOI: 10.1016/S0140-6736(12)60681-3. PMID: 22763110. PMCID: PMC3443899.15. Burgess S, Butterworth A, Thompson SG. 2013; Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 37:658–65. DOI: 10.1002/gepi.21758. PMID: 24114802. PMCID: PMC4377079.
Article16. Pierce BL, Burgess S. 2013; Efficient design for Mendelian randomization studies: subsample and 2-sample instrumental variable estimators. Am J Epidemiol. 178:1177–84. DOI: 10.1093/aje/kwt084. PMID: 23863760. PMCID: PMC3783091.
Article17. Hartwig FP, Davies NM, Hemani G, Davey Smith G. 2016; Two-sample Mendelian randomization: avoiding the downsides of a powerful, widely applicable but potentially fallible technique. Int J Epidemiol. 45:1717–26. DOI: 10.1093/ije/dyx028. PMID: 28338968. PMCID: PMC5722032.
Article18. Bowden J, Davey Smith G, Burgess S. 2015; Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 44:512–25. DOI: 10.1093/ije/dyv080. PMID: 26050253. PMCID: PMC4469799.
Article19. Burgess S, Thompson SG. 2017; Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol. 32:377–89. DOI: 10.1007/s10654-017-0255-x. PMID: 28527048. PMCID: PMC5506233.
Article20. Bowden J, Davey Smith G, Haycock PC, Burgess S. 2016; Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 40:304–14. DOI: 10.1002/gepi.21965. PMID: 27061298. PMCID: PMC4849733.
Article21. Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, et al. 2018; The MR-Base platform supports systematic causal inference across the human phenome. Elife. 7:e34408. DOI: 10.7554/eLife.34408. PMID: 29846171. PMCID: PMC5976434.
Article22. Egger M, Smith GD, Phillips AN. 1997; Meta-analysis: principles and procedures. BMJ. 315:1533–7. DOI: 10.1136/bmj.315.7121.1533. PMID: 9432252. PMCID: PMC2127925.
Article23. Courties A, Sellam J, Berenbaum F. 2017; Metabolic syndrome- associated osteoarthritis. Curr Opin Rheumatol. 29:214–22. DOI: 10.1097/BOR.0000000000000373. PMID: 28072592.24. Schott EM, Farnsworth CW, Grier A, Lillis JA, Soniwala S, Dadourian GH, et al. 2018; Targeting the gut microbiome to treat the osteoarthritis of obesity. JCI Insight. 3:e95997. DOI: 10.1172/jci.insight.95997. PMID: 29669931. PMCID: PMC5931133.
Article25. Metcalfe D, Harte AL, Aletrari MO, Al Daghri NM, Al Disi D, Tripathi G, et al. 2012; Does endotoxaemia contribute to osteoarthritis in obese patients? Clin Sci (Lond). 123:627–34. DOI: 10.1042/CS20120073. PMID: 22888972.
Article26. Thompson JR, Minelli C, Bowden J, Del Greco FM, Gill D, Jones EM, et al. 2017; Mendelian randomization incorporating uncertainty about pleiotropy. Stat Med. 36:4627–45. DOI: 10.1002/sim.7442. PMID: 28850703.
Article27. Smith GD, Ebrahim S. 2004; Mendelian randomization: prospects, potentials, and limitations. Int J Epidemiol. 33:30–42. DOI: 10.1093/ije/dyh132. PMID: 15075143.
Article28. Swerdlow DI, Kuchenbaecker KB, Shah S, Sofat R, Holmes MV, White J, et al. 2016; Selecting instruments for Mendelian randomization in the wake of genome-wide association studies. Int J Epidemiol. 45:1600–16. DOI: 10.1093/ije/dyw088. PMID: 27342221. PMCID: PMC5100611.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- The Uric Acid and Gout have No Direct Causality With Osteoarthritis: A Mendelian Randomization Study
- Causal Association between Bone Mineral Density and Osteoarthritis: A Mendelian Randomization Study
- The Association between the Gut Microbiota and Erectile Dysfunction
- Causal association between serum bilirubin and ischemic stroke: multivariable Mendelian randomization
- Complex influences of gut microbiome metabolism on various drug responses