Int J Stem Cells.
2023 Aug;16(3):342-355. 10.15283/ijsc22101.
Dissection of Cellular Communication between Human Primary Osteoblasts and Bone Marrow Mesenchymal Stem Cells in Osteoarthritis at Single-Cell Resolution
- Affiliations
-
- 1Laboratory of Molecular and Statistical Genetics, College of Life Sciences, Hunan Normal University, Changsha, China
- 2Tulane Center of Biomedical Informatics and Genomics, Deming Department of Medicine, Tulane University School of Medicine, New Orleans, LA, USA
- 3School of Basic Medical Science, Central South University, Changsha, China
- 4Department of Orthopedics, Xiangya Hospital, Central South University, Changsha, China
- 5Department of Orthopedics and National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Central South University, Changsha, China
- 6Center of Reproductive Health, System Biology and Data Information, Institute of Reproductive & Stem Cell Engineering, School of Basic Medical Science, Central South University, Changsha, China
- KMID: 2545233
- DOI: http://doi.org/10.15283/ijsc22101
Abstract
- Background and Objectives
Osteoblasts are derived from bone marrow mesenchymal stem cells (BMMSCs) and play important role in bone remodeling. While our previous studies have investigated the cell subtypes and heterogeneity in osteoblasts and BMMSCs separately, cell-to-cell communications between osteoblasts and BMMSCs in vivo in humans have not been characterized. The aim of this study was to investigate the cellular communication between human primary osteoblasts and bone marrow mesenchymal stem cells.
Methods and Results
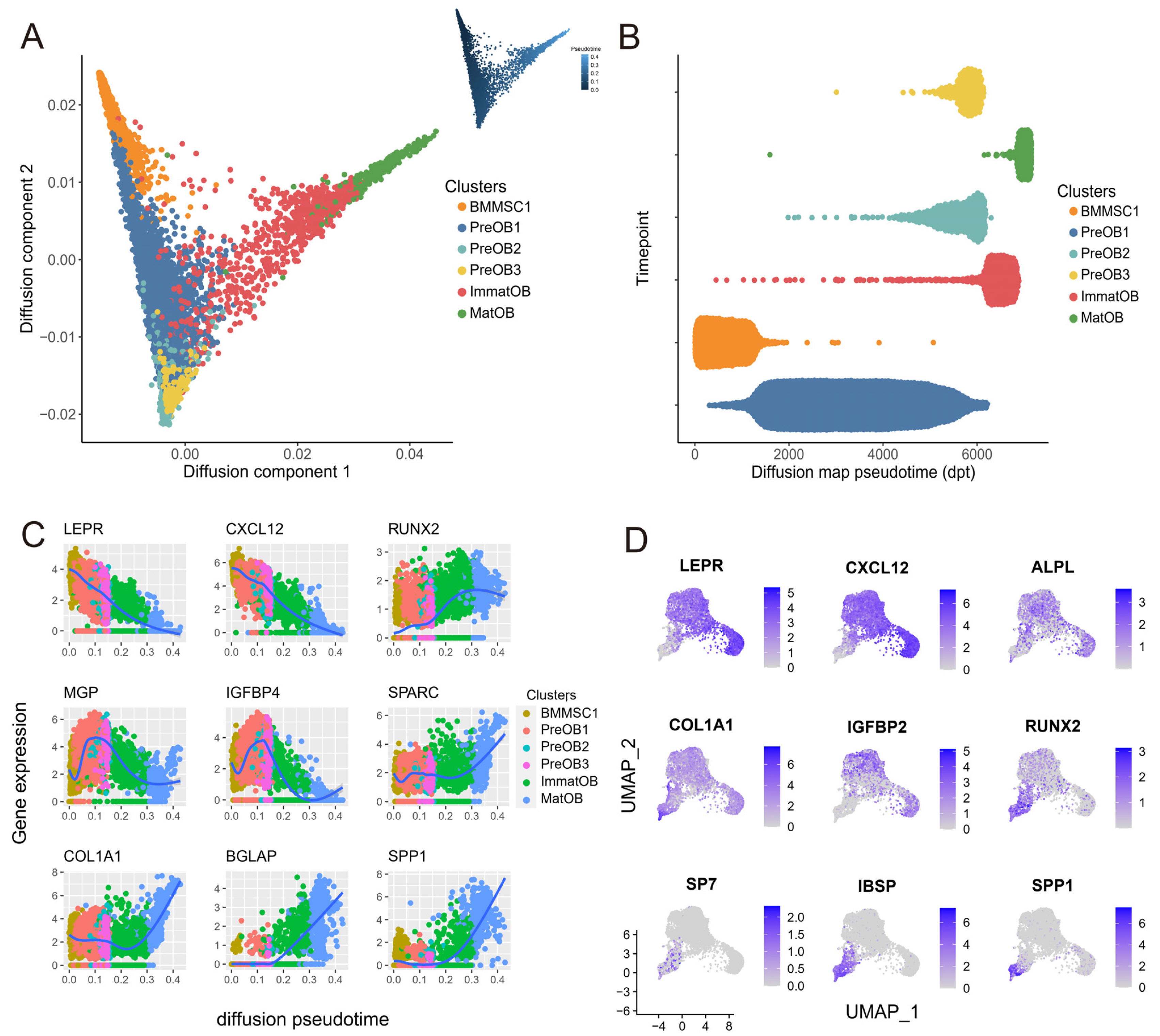
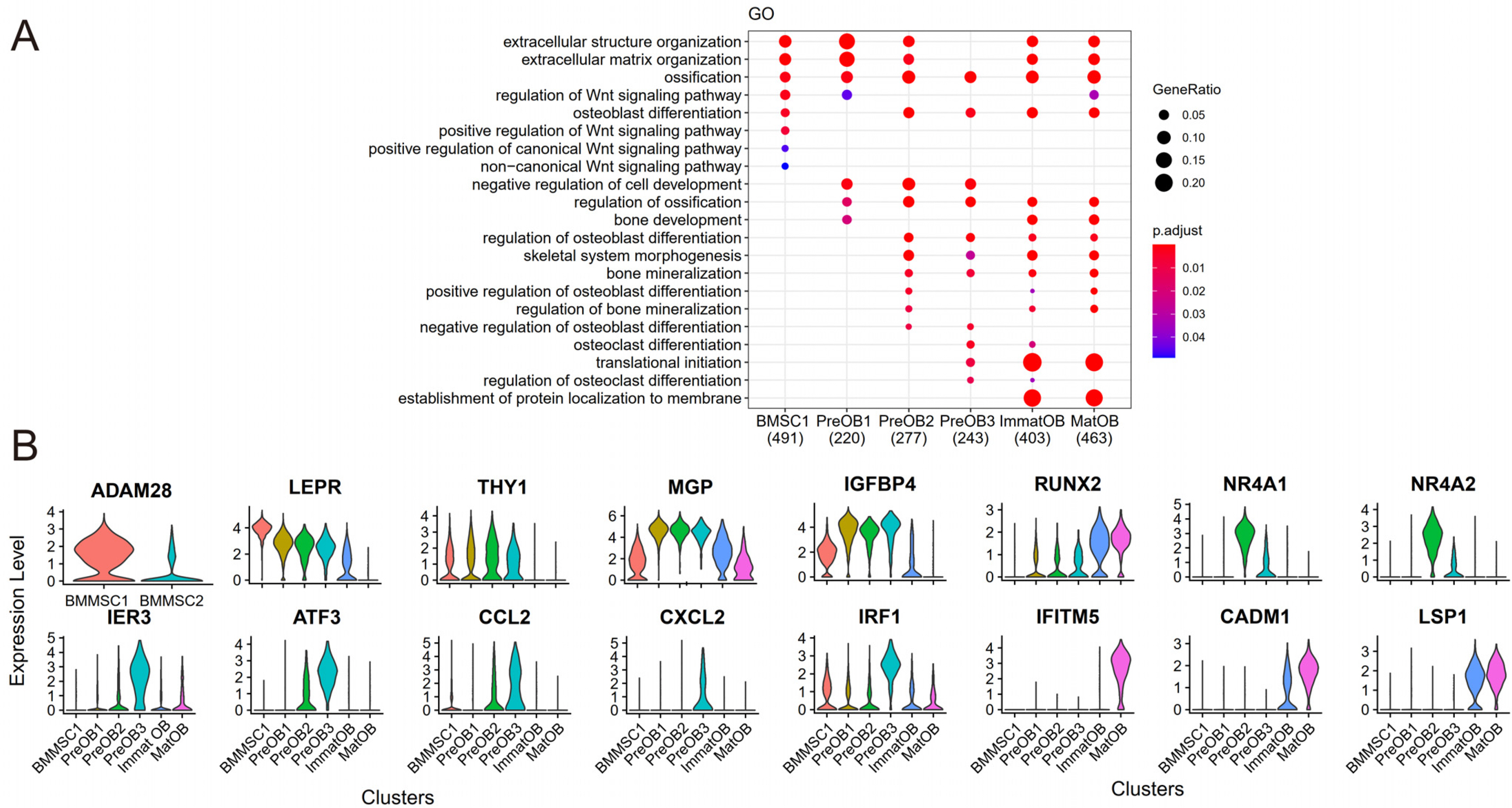
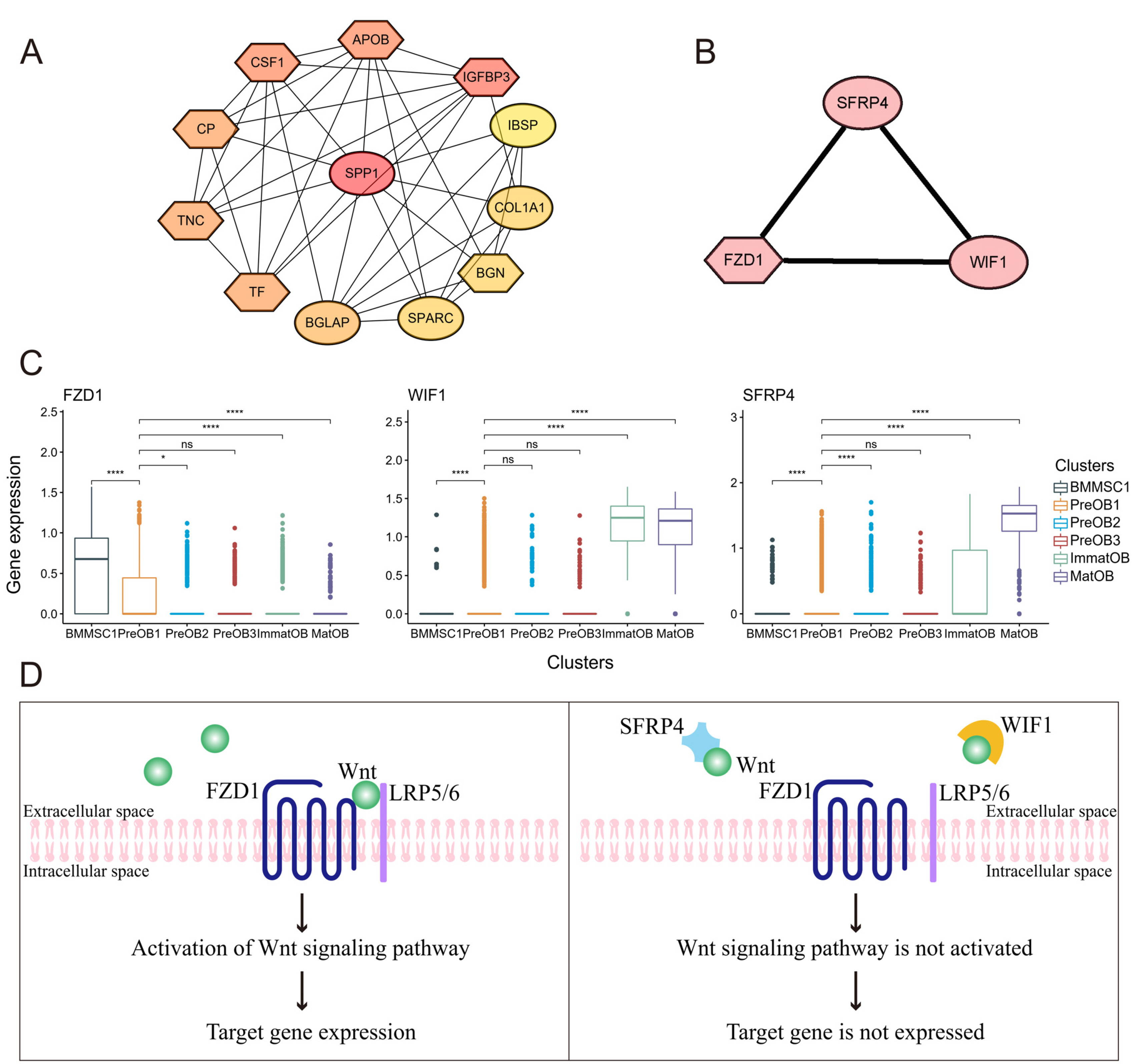
To investigate the cell-to-cell communications between osteoblasts and BMMSCs and identify new cell subtypes, we performed a systematic integration analysis with our single-cell RNA sequencing (scRNA-seq) transcriptomes data from BMMSCs and osteoblasts. We successfully identified a novel preosteoblasts subtype which highly expressed ATF3, CCL2, CXCL2 and IRF1. Biological functional annotations of the transcriptomes suggested that the novel preosteoblasts subtype may inhibit osteoblasts differentiation, maintain cells to a less differentiated status and recruit osteoclasts. Ligand-receptor interaction analysis showed strong interaction between mature osteoblasts and BMMSCs. Meanwhile, we found FZD1 was highly expressed in BMMSCs of osteogenic differentiation direction. WIF1 and SFRP4, which were highly expressed in mature osteoblasts were reported to inhibit osteogenic differentiation. We speculated that WIF1 and sFRP4 expressed in mature osteoblasts inhibited the binding of FZD1 to Wnt ligand in BMMSCs, thereby further inhibiting osteogenic differentiation of BMMSCs.
Conclusions
Our study provided a more systematic and comprehensive understanding of the heterogeneity of osteogenic cells. At the single cell level, this study provided insights into the cell-to-cell communications between BMMSCs and osteoblasts and mature osteoblasts may mediate negative feedback regulation of osteogenesis process.
Keyword
Figure
Reference
-
References
1. Martin TJ, Seeman E. 2008; Bone remodelling: its local regulation and the emergence of bone fragility. Best Pract Res Clin Endocrinol Metab. 22:701–722. DOI: 10.1016/j.beem.2008.07.006. PMID: 19028353.
Article2. Muruganandan S, Sinal CJ. 2014; The impact of bone marrow adipocytes on osteoblast and osteoclast differentiation. IUBMB Life. 66:147–155. DOI: 10.1002/iub.1254. PMID: 24638917.
Article3. Chen Q, Shou P, Zheng C, Jiang M, Cao G, Yang Q, Cao J, Xie N, Velletri T, Zhang X, Xu C, Zhang L, Yang H, Hou J, Wang Y, Shi Y. 2016; Fate decision of mesenchymal stem cells: adipocytes or osteoblasts? Cell Death Differ. 23:1128–1139. DOI: 10.1038/cdd.2015.168. PMID: 26868907. PMCID: PMC4946886.
Article4. Zieba JT, Chen YT, Lee BH, Bae Y. 2020; Notch signaling in skeletal development, homeostasis and pathogenesis. Bio-molecules. 10:332. DOI: 10.3390/biom10020332. PMID: 32092942. PMCID: PMC7072615.
Article5. Maeda K, Kobayashi Y, Koide M, Uehara S, Okamoto M, Ishihara A, Kayama T, Saito M, Marumo K. 2019; The regulation of bone metabolism and disorders by Wnt signaling. Int J Mol Sci. 20:5525. DOI: 10.3390/ijms20225525. PMID: 31698687. PMCID: PMC6888566.
Article6. Liu S, Stroncek DF, Zhao Y, Chen V, Shi R, Chen J, Ren J, Liu H, Bae HJ, Highfill SL, Jin P. 2019; Single cell sequencing reveals gene expression signatures associated with bone marrow stromal cell subpopulations and time in culture. J Transl Med. 17:23. DOI: 10.1186/s12967-018-1766-2. PMID: 30635013. PMCID: PMC6330466.
Article7. Wang Z, Li X, Yang J, Gong Y, Zhang H, Qiu X, Liu Y, Zhou C, Chen Y, Greenbaum J, Cheng L, Hu Y, Xie J, Yang X, Li Y, Schiller MR, Chen Y, Tan L, Tang SY, Shen H, Xiao HM, Deng HW. 2021; Single-cell RNA sequencing deconvolutes the in vivo heterogeneity of human bone marrow-derived mesenchymal stem cells. Int J Biol Sci. 17:4192–4206. DOI: 10.7150/ijbs.61950. PMID: 34803492. PMCID: PMC8579438.
Article8. Gong Y, Yang J, Li X, Zhou C, Chen Y, Wang Z, Qiu X, Liu Y, Zhang H, Greenbaum J, Cheng L, Hu Y, Xie J, Yang X, Li Y, Bai Y, Wang YP, Chen Y, Tan LJ, Shen H, Xiao HM, Deng HW. 2021; A systematic dissection of human primary osteoblasts in vivo at single-cell resolution. Aging (Albany NY). 13:20629–20650. DOI: 10.18632/aging.203452. PMID: 34428745. PMCID: PMC8436943.
Article9. Fujita K, Roforth MM, Atkinson EJ, Peterson JM, Drake MT, McCready LK, Farr JN, Monroe DG, Khosla S. 2014; Isolation and characterization of human osteoblasts from needle biopsies without in vitro culture. Osteoporos Int. 25:887–895. DOI: 10.1007/s00198-013-2529-9. PMID: 24114401. PMCID: PMC4216562.
Article10. Quirici N, Soligo D, Bossolasco P, Servida F, Lumini C, Deliliers GL. 2002; Isolation of bone marrow mesenchymal stem cells by anti-nerve growth factor receptor antibodies. Exp Hematol. 30:783–791. DOI: 10.1016/S0301-472X(02)00812-3. PMID: 12135677.
Article11. Poloni A, Maurizi G, Rosini V, Mondini E, Mancini S, Discepoli G, Biasio S, Battaglini G, Felicetti S, Berardinelli E, Serrani F, Leoni P. 2009; Selection of CD271(+) cells and human AB serum allows a large expansion of mesenchymal stromal cells from human bone marrow. Cytotherapy. 11:153–162. DOI: 10.1080/14653240802582125. PMID: 19301169.
Article12. Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. 2018; Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 36:411–420. DOI: 10.1038/nbt.4096. PMID: 29608179. PMCID: PMC6700744.
Article13. Brun J, Lutz KA, Neumayer KM, Klein G, Seeger T, Uynuk-Ool T, Wörgötter K, Schmid S, Kraushaar U, Guenther E, Rolauffs B, Aicher WK, Hart ML. 2015; Smooth muscle-like cells generated from human mesenchymal stromal cells display marker gene expression and electrophysiological competence comparable to bladder smooth muscle cells. PLoS One. 10:e0145153. DOI: 10.1371/journal.pone.0145153. PMID: 26673782. PMCID: PMC4684225.
Article14. Angerer P, Haghverdi L, Büttner M, Theis FJ, Marr C, Buettner F. 2016; destiny: diffusion maps for large-scale single-cell data in R. Bioinformatics. 32:1241–1243. DOI: 10.1093/bioinformatics/btv715. PMID: 26668002.15. Wang Y, Wang R, Zhang S, Song S, Jiang C, Han G, Wang M, Ajani J, Futreal A, Wang L. 2019; iTALK: an R package to characterize and illustrate intercellular communication. bioRxiv. doi:10.1101/507871. DOI: 10.1101/507871.
Article16. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003; Cytoscape: a software environment for integrated models of biomo-lecular interaction networks. Genome Res. 13:2498–2504. DOI: 10.1101/gr.1239303. PMID: 14597658. PMCID: PMC403769.
Article17. Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. 2014; cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 8(Suppl 4):S11. DOI: 10.1186/1752-0509-8-S4-S11. PMID: 25521941. PMCID: PMC4290687.
Article18. Becht E, McInnes L, Healy J, Dutertre CA, Kwok IWH, Ng LG, Ginhoux F, Newell EW. 2018; Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biote-chnol. doi: 10.1038/nbt.4314 [Epub ahead of print]. DOI: 10.1038/nbt.4314. PMID: 30531897.
Article19. Rutkovskiy A, Stensløkken KO, Vaage IJ. 2016; Osteoblast differentiation at a glance. Med Sci Monit Basic Res. 22:95–106. DOI: 10.12659/MSMBR.901142. PMID: 27667570. PMCID: PMC5040224.
Article20. Zhao Z, Wen LY, Jin M, Deng ZH, Jin Y. 2006; ADAM28 participates in the regulation of tooth development. Arch Oral Biol. 51:996–1005. DOI: 10.1016/j.archoralbio.2006.05.010. PMID: 16836973.
Article21. Zhao Z, Tang L, Deng Z, Wen L, Jin Y. 2008; Essential role of ADAM28 in regulating the proliferation and differentiation of human dental papilla mesenchymal cells (hDPMCs). Histochem Cell Biol. 130:1015–1025. DOI: 10.1007/s00418-008-0467-y. PMID: 18690470.
Article22. Zhang J, Ma Z, Yan K, Wang Y, Yang Y, Wu X. 2019; Matrix Gla protein promotes the bone formation by up-regulating Wnt/β-catenin signaling pathway. Front Endocrinol (Lausanne). 10:891. DOI: 10.3389/fendo.2019.00891. PMID: 31920993. PMCID: PMC6933527.
Article23. Li H, Yu S, Hao F, Sun X, Zhao J, Xu Q, Duan D. 2018; Insulin-like growth factor binding protein 4 inhibits proliferation of bone marrow mesenchymal stem cells and enhances growth of neurospheres derived from the stem cells. Cell Biochem Funct. 36:331–341. DOI: 10.1002/cbf.3353. PMID: 30028031.
Article24. Wu J, Wang C, Miao X, Wu Y, Yuan J, Ding M, Li J, Shi Z. 2017; Age-related insulin-like growth factor binding protein-4 overexpression inhibits osteogenic differentiation of rat mesenchymal stem cells. Cell Physiol Biochem. 42:640–650. DOI: 10.1159/000477873. PMID: 28595186.
Article25. Li X, Wei W, Huynh H, Zuo H, Wang X, Wan Y. 2015; Nur77 prevents excessive osteoclastogenesis by inducing ubiquitin ligase Cbl-b to mediate NFATc1 self-limitation. Elife. 4:e07217. DOI: 10.7554/eLife.07217. PMID: 26173181. PMCID: PMC4518709.
Article26. Lee MK, Choi H, Gil M, Nikodem VM. 2006; Regulation of osteoblast differentiation by Nurr1 in MC3T3-E1 cell line and mouse calvarial osteoblasts. J Cell Biochem. 99:986–994. DOI: 10.1002/jcb.20990. PMID: 16741951.
Article27. Salem S, Gao C, Li A, Wang H, Nguyen-Yamamoto L, Goltzman D, Henderson JE, Gros P. 2014; A novel role for interferon regulatory factor 1 (IRF1) in regulation of bone metabolism. J Cell Mol Med. 18:1588–1598. DOI: 10.1111/jcmm.12327. PMID: 24954358. PMCID: PMC4152406.
Article28. Park JK, Jang H, Hwang S, Kim EJ, Kim DE, Oh KB, Kwon DJ, Koh JT, Kimura K, Inoue H, Jang WG, Lee JW. 2014; ER stress-inducible ATF3 suppresses BMP2-induced ALP expression and activation in MC3T3-E1 cells. Biochem Biophys Res Commun. 443:333–338. DOI: 10.1016/j.bbrc.2013.11.121. PMID: 24315873.
Article29. Siddiqui JA, Partridge NC. 2017; CCL2/monocyte chemoattra-ctant protein 1 and parathyroid hormone action on bone. Front Endocrinol (Lausanne). 8:49. DOI: 10.3389/fendo.2017.00049. PMID: 28424660. PMCID: PMC5372820.
Article30. Yang Y, Zhou X, Li Y, Chen A, Liang W, Liang G, Huang B, Li Q, Jin D. 2019; CXCL2 attenuates osteoblast differentiation by inhibiting the ERK1/2 signaling pathway. J Cell Sci. 132:jcs230490. DOI: 10.1242/jcs.230490. PMID: 31292171.31. Ha J, Lee Y, Kim HH. 2011; CXCL2 mediates lipopolysaccharide-induced osteoclastogenesis in RANKL-primed precursors. Cytokine. 55:48–55. DOI: 10.1016/j.cyto.2011.03.026. PMID: 21507677.
Article32. Arlt A, Schäfer H. 2011; Role of the immediate early response 3 (IER3) gene in cellular stress response, inflammation and tumorigenesis. Eur J Cell Biol. 90:545–552. DOI: 10.1016/j.ejcb.2010.10.002. PMID: 21112119.
Article33. Semler O, Garbes L, Keupp K, Swan D, Zimmermann K, Becker J, Iden S, Wirth B, Eysel P, Koerber F, Schoenau E, Bohlander SK, Wollnik B, Netzer C. 2012; A mutation in the 5'-UTR of IFITM5 creates an in-frame start codon and causes autosomal-dominant osteogenesis imperfecta type V with hyperplastic callus. Am J Hum Genet. 91:349–357. DOI: 10.1016/j.ajhg.2012.06.011. PMID: 22863195. PMCID: PMC3415541.
Article34. Inoue T, Hagiyama M, Enoki E, Sakurai MA, Tan A, Wakayama T, Iseki S, Murakami Y, Fukuda K, Hamanishi C, Ito A. 2013; Cell adhesion molecule 1 is a new osteoblastic cell adhesion molecule and a diagnostic marker for osteosarcoma. Life Sci. 92:91–99. DOI: 10.1016/j.lfs.2012.10.021. PMID: 23142238.
Article35. Pulford K, Jones M, Banham AH, Haralambieva E, Mason DY. 1999; Lymphocyte-specific protein 1: a specific marker of human leucocytes. Immunology. 96:262–271. DOI: 10.1046/j.1365-2567.1999.00677.x. PMID: 10233704. PMCID: PMC2326732.
Article36. Liu L, Cara DC, Kaur J, Raharjo E, Mullaly SC, Jongstra-Bilen J, Jongstra J, Kubes P. 2005; LSP1 is an endothelial gatekeeper of leukocyte transendothelial migration. J Exp Med. 201:409–418. DOI: 10.1084/jem.20040830. PMID: 15684321. PMCID: PMC2213033.
Article37. Campbell ID, Humphries MJ. 2011; Integrin structure, activation, and interactions. Cold Spring Harb Perspect Biol. 3:a004994. DOI: 10.1101/cshperspect.a004994. PMID: 21421922. PMCID: PMC3039929.
Article38. Marie PJ. 2013; Targeting integrins to promote bone formation and repair. Nat Rev Endocrinol. 9:288–295. DOI: 10.1038/nrendo.2013.4. PMID: 23358353.
Article39. Chen Q, Shou P, Zhang L, Xu C, Zheng C, Han Y, Li W, Huang Y, Zhang X, Shao C, Roberts AI, Rabson AB, Ren G, Zhang Y, Wang Y, Denhardt DT, Shi Y. 2014; An osteopontin-integrin interaction plays a critical role in directing adipogenesis and osteogenesis by mesenchymal stem cells. Stem Cells. 32:327–337. DOI: 10.1002/stem.1567. PMID: 24123709. PMCID: PMC3961005.
Article40. Reyes CD, García AJ. 2004; Alpha2beta1 integrin-specific collagen-mimetic surfaces supporting osteoblastic differen-tiation. J Biomed Mater Res A. 69:591–600. DOI: 10.1002/jbm.a.30034. PMID: 15162400.41. Gronthos S, Simmons PJ, Graves SE, Robey PG. 2001; Integrin-mediated interactions between human bone marrow stromal precursor cells and the extracellular matrix. Bone. 28:174–181. DOI: 10.1016/S8756-3282(00)00424-5. PMID: 11182375.
Article42. Florencio-Silva R, Sasso GR, Sasso-Cerri E, Simões MJ, Cerri PS. 2015; Biology of bone tissue: structure, function, and factors that influence bone cells. Biomed Res Int. 2015:421746. DOI: 10.1155/2015/421746. PMID: 26247020. PMCID: PMC4515490.
Article43. Fu HL, Valiathan RR, Arkwright R, Sohail A, Mihai C, Kumarasiri M, Mahasenan KV, Mobashery S, Huang P, Agarwal G, Fridman R. 2013; Discoidin domain receptors: unique receptor tyrosine kinases in collagen-mediated signaling. J Biol Chem. 288:7430–7437. DOI: 10.1074/jbc.R112.444158. PMID: 23335507. PMCID: PMC3597784.
Article44. Yang H, Sun L, Cai W, Gu J, Xu D, Deb A, Duan J. 2020; DDR2, a discoidin domain receptor, is a marker of periosteal osteoblast and osteoblast progenitors. J Bone Miner Metab. 38:670–677. DOI: 10.1007/s00774-020-01108-y. PMID: 32415375. PMCID: PMC7581459.
Article45. Ruiz PA, Jarai G. 2011; Collagen I induces discoidin domain receptor (DDR) 1 expression through DDR2 and a JAK2-ERK1/2-mediated mechanism in primary human lung fibroblasts. J Biol Chem. 286:12912–12923. DOI: 10.1074/jbc.M110.143693. PMID: 21335558. PMCID: PMC3075638.
Article46. Yu S, Yerges-Armstrong LM, Chu Y, Zmuda JM, Zhang Y. 2015; AP2 suppresses osteoblast differentiation and mineralization through down-regulation of Frizzled-1. Biochem J. 465:395–404. DOI: 10.1042/BJ20140668. PMID: 25369469.
Article47. Yu S, Yerges-Armstrong LM, Chu Y, Zmuda JM, Zhang Y. 2013; E2F1 effects on osteoblast differentiation and mineralization are mediated through up-regulation of frizzled-1. Bone. 56:234–241. DOI: 10.1016/j.bone.2013.06.019. PMID: 23806799. PMCID: PMC3758927.
Article48. Katoh Y, Katoh M. 2005; Comparative genomics on Dkk1 orthologs. Int J Oncol. 27:275–279. DOI: 10.3892/ijo.27.1.275. PMID: 15942669.
Article49. Pez F, Lopez A, Kim M, Wands JR, Caron de Fromentel C, Merle P. 2013; Wnt signaling and hepatocarcinogenesis: molecular targets for the development of innovative anticancer drugs. J Hepatol. 59:1107–1117. DOI: 10.1016/j.jhep.2013.07.001. PMID: 23835194.
Article50. Nakanishi R, Shimizu M, Mori M, Akiyama H, Okudaira S, Otsuki B, Hashimoto M, Higuchi K, Hosokawa M, Tsuboyama T, Nakamura T. 2006; Secreted frizzled-related protein 4 is a negative regulator of peak BMD in SAMP6 mice. J Bone Miner Res. 21:1713–1721. DOI: 10.1359/jbmr.060719. PMID: 17002585.
Article51. Nakanishi R, Akiyama H, Kimura H, Otsuki B, Shimizu M, Tsuboyama T, Nakamura T. 2008; Osteoblast-targeted expression of Sfrp4 in mice results in low bone mass. J Bone Miner Res. 23:271–277. DOI: 10.1359/jbmr.071007. PMID: 17907918.
Article52. Yamada A, Iwata T, Yamato M, Okano T, Izumi Y. 2013; Diverse functions of secreted frizzled-related proteins in the osteoblastogenesis of human multipotent mesenchymal stromal cells. Biomaterials. 34:3270–3278. DOI: 10.1016/j.biomaterials.2013.01.066. PMID: 23384792.
Article53. Baker EK, Taylor S, Gupte A, Chalk AM, Bhattacharya S, Green AC, Martin TJ, Strbenac D, Robinson MD, Purton LE, Walkley CR. 2015; Wnt inhibitory factor 1 (WIF1) is a marker of osteoblastic differentiation stage and is not silenced by DNA methylation in osteosarcoma. Bone. 73:223–232. DOI: 10.1016/j.bone.2014.12.063. PMID: 25571841.
Article54. Kansara M, Tsang M, Kodjabachian L, Sims NA, Trivett MK, Ehrich M, Dobrovic A, Slavin J, Choong PF, Simmons PJ, Dawid IB, Thomas DM. 2009; Wnt inhibitory factor 1 is epigenetically silenced in human osteosarcoma, and targeted disruption accelerates osteosarcomagenesis in mice. J Clin Invest. 119:837–851. DOI: 10.1172/JCI37175. PMID: 19307728. PMCID: PMC2662557.
Article55. Cho SW, Yang JY, Sun HJ, Jung JY, Her SJ, Cho HY, Choi HJ, Kim SW, Kim SY, Shin CS. 2009; Wnt inhibitory factor (WIF)-1 inhibits osteoblastic differentiation in mouse embryonic mesenchymal cells. Bone. 44:1069–1077. DOI: 10.1016/j.bone.2009.02.012. PMID: 19254785.56. Yue R, Zhou BO, Shimada IS, Zhao Z, Morrison SJ. 2016; Leptin receptor promotes adipogenesis and reduces osteogenesis by regulating mesenchymal stromal cells in adult bone marrow. Cell Stem Cell. 18:782–796. DOI: 10.1016/j.stem.2016.02.015. PMID: 27053299.57. Aubin JE. 2001; Regulation of osteoblast formation and function. Rev Endocr Metab Disord. 2:81–94. DOI: 10.1023/A:1010011209064. PMID: 11704982.58. Komori T. 2006; Regulation of osteoblast differentiation by transcription factors. J Cell Biochem. 99:1233–1239. DOI: 10.1002/jcb.20958. PMID: 16795049.59. Matic I, Matthews BG, Wang X, Dyment NA, Worthley DL, Rowe DW, Grcevic D, Kalajzic I. 2016; Quiescent bone lining cells are a major source of osteoblasts during adulthood. Stem Cells. 34:2930–2942. DOI: 10.1002/stem.2474. PMID: 27507737. PMCID: PMC5450652.60. Mikami Y, Yamamoto K, Akiyama Y, Kobayashi M, Watanabe E, Watanabe N, Asano M, Shimizu N, Komiyama K. 2015; Osteogenic gene transcription is regulated via gap junction-mediated cell-cell communication. Stem Cells Dev. 24:214–227. DOI: 10.1089/scd.2014.0060. PMID: 25137151. PMCID: PMC4291207.61. Yang X, Yang J, Lei P, Wen T. 2019; LncRNA MALAT1 shuttled by bone marrow-derived mesenchymal stem cells-secreted exosomes alleviates osteoporosis through mediating micro-RNA-34c/SATB2 axis. Aging (Albany NY). 11:8777–8791. DOI: 10.18632/aging.102264. PMID: 31659145. PMCID: PMC6834402.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Interaction of Mesenchymal Stem Cells and Osteoblasts for in vitro Osteogenesis
- Clinical Safety and Efficacy of Autologous Bone Marrow-Derived Mesenchymal Stem Cell Transplantation in Sensorineural Hearing Loss Patients
- Osteoblastic differentiation of adult stem cells by Biphasic Calcium Phosphate
- Stem Cells and Niemann Pick Disease
- Clinical Use of Mesenchymal Stem Cells in Bone Regeneration