J Pathol Transl Med.
2022 Nov;56(6):326-333. 10.4132/jptm.2022.10.17.
Biomarker testing of cytology specimens in personalized medicine for lung cancer patients
- Affiliations
-
- 1Department of Pathology and Translational Medicine, Seoul National University Bundang Hospital, Seongnam, Korea
- 2Department of Pathology, Seoul National University College of Medicine, Seoul, Korea
- KMID: 2535852
- DOI: http://doi.org/10.4132/jptm.2022.10.17
Abstract
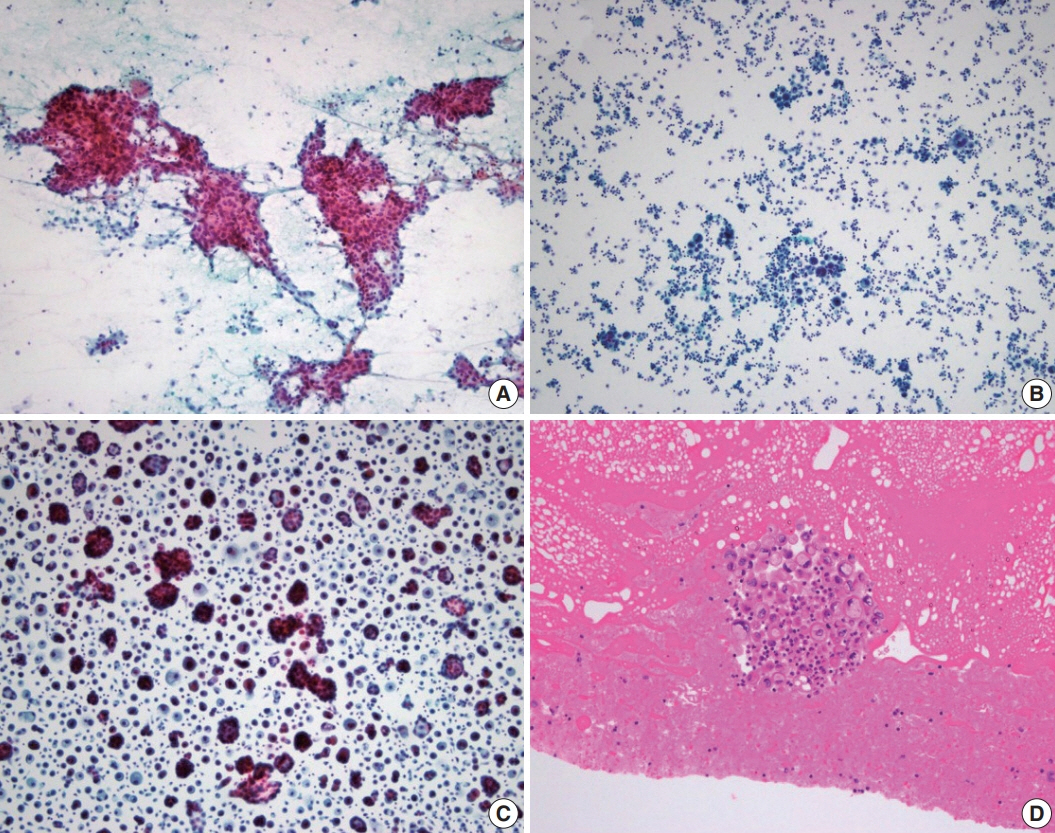
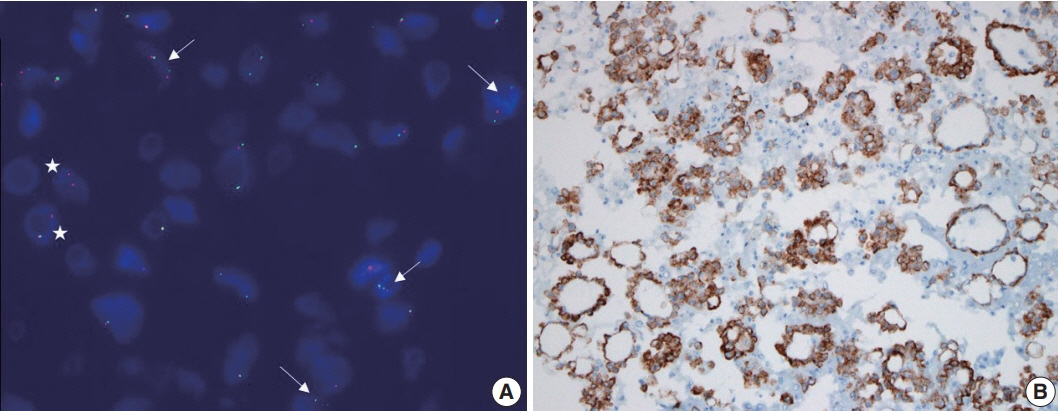
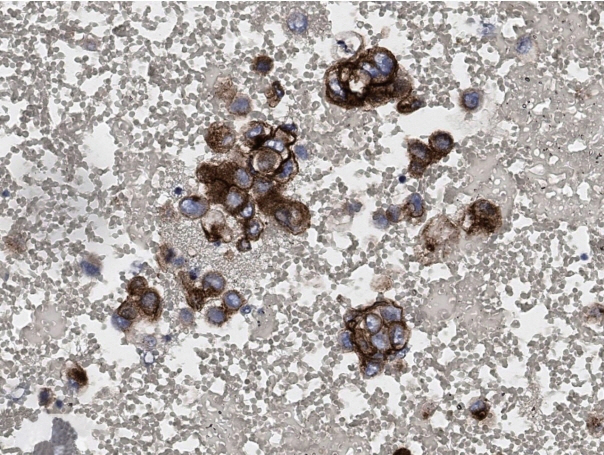
- Every patient with advanced non–small cell lung cancer (NSCLC) should be tested for targetable driver mutations and gene arrangements that may open avenues for targeted therapy. As most patients with NSCLC in the advanced stage of the disease are not candidates for surgery, these tests have to be performed on small biopsies or cytology samples. A growing number of other genetic changes with targetable mutations may be treatable in the near future. To identify patients who might benefit from novel targeted therapy, relevant markers should be tested in an appropriate context. In addition, immunotherapy of lung cancer is guided by the status of programmed death-ligand 1 expression in tumor cells. The variety and versatility of cytological specimen preparations offer significant advantages for molecular testing; however, they frequently remain underused. Therefore, evaluating the utility and adequacy of cytologic specimens is important, not only from a lung cancer diagnosis, but also for the large number of ancillary studies that are necessary to provide appropriate clinical management. A large proportion of lung cancers is diagnosed by aspiration or exfoliative cytology specimens; thus, optimizing strategies to triage and best use the tissue for diagnosis and biomarker studies forms a critical component of lung cancer management. In this review, we discuss the opportunities and challenges of using cytologic specimens for biomarker testing of lung cancer and the role of cytopathology in the molecular era.
Keyword
Figure
Cited by 1 articles
-
Next step of molecular pathology: next-generation sequencing in cytology
Ricella Souza da Silva, Fernando Schmitt
J Pathol Transl Med. 2024;58(6):291-298. doi: 10.4132/jptm.2024.10.22.
Reference
-
References
1. Lindeman NI, Cagle PT, Beasley MB, et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. J Thorac Oncol. 2013; 8:823–59.
Article2. Lindeman NI, Cagle PT, Aisner DL, et al. Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: guideline from the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. J Thorac Oncol. 2018; 13:323–58.3. Camidge DR, Doebele RC, Kerr KM. Comparing and contrasting predictive biomarkers for immunotherapy and targeted therapy of NSCLC. Nat Rev Clin Oncol. 2019; 16:341–55.
Article4. Roh MH. The Utilization of cytologic fine-needle aspirates of lung cancer for molecular diagnostic testing. J Pathol Transl Med. 2015; 49:300–9.
Article5. Roy-Chowdhuri S, Stewart J. Preanalytic variables in cytology: lessons learned from next-generation sequencing: the MD Anderson experience. Arch Pathol Lab Med. 2016; 140:1191–9.6. Hwang DH, Garcia EP, Ducar MD, Cibas ES, Sholl LM. Next-generation sequencing of cytologic preparations: an analysis of quality metrics. Cancer Cytopathol. 2017; 125:786–94.
Article7. Harada S, Agosto-Arroyo E, Levesque JA, et al. Poor cell block adequacy rate for molecular testing improved with the addition of DiffQuik-stained smears: need for better cell block processing. Cancer Cytopathol. 2015; 123:480–7.
Article8. Velizheva NP, Rechsteiner MP, Wong CE, et al. Cytology smears as excellent starting material for next-generation sequencing-based molecular testing of patients with adenocarcinoma of the lung. Cancer Cytopathol. 2017; 125:30–40.
Article9. Huang M, Wei S. Overview of molecular testing of cytology specimens. Acta Cytol. 2020; 64:136–46.
Article10. Petriella D, Galetta D, Rubini V, et al. Molecular profiling of thinprep FNA samples in assisting clinical management of non-smallcell lung cancer. Mol Biotechnol. 2013; 54:913–9.
Article11. Abedi-Ardekani B, Vielh P. Is liquid-based cytology the magic bullet for performing molecular techniques? Acta Cytol. 2014; 58:574–81.
Article12. Zeppa P. Liquid-based cytology: a 25-year bridge between the pap smear and molecular cytopathology. Acta Cytol. 2014; 58:519–21.
Article13. Bellevicine C, Malapelle U, Vigliar E, de Luca C, Troncone G. Epidermal growth factor receptor test performed on liquid-based cytology lung samples: experience of an academic referral center. Acta Cytol. 2014; 58:589–94.
Article14. Malapelle U, de Rosa N, Bellevicine C, et al. EGFR mutations detection on liquid-based cytology: is microscopy still necessary? J Clin Pathol. 2012; 65:561–4.
Article15. Reynolds JP, Tubbs RR, Minca EC, et al. EGFR mutational genotyping of liquid based cytology samples obtained via fine needle aspiration (FNA) at endobronchial ultrasound of non-small cell lung cancer (NSCLC). Lung Cancer. 2014; 86:158–63.
Article16. Dejmek A, Zendehrokh N, Tomaszewska M, Edsjo A. Preparation of DNA from cytological material: effects of fixation, staining, and mounting medium on DNA yield and quality. Cancer Cytopathol. 2013; 121:344–53.17. Kim WY, Oh SY, Kim H, Hwang TS. DNA degradation in liquidbased cytology and its comparison with conventional smear. Diagn Cytopathol. 2016; 44:450–8.
Article18. Lozano MD, Zulueta JJ, Echeveste JI, et al. Assessment of epidermal growth factor receptor and K-ras mutation status in cytological stained smears of non-small cell lung cancer patients: correlation with clinical outcomes. Oncologist. 2011; 16:877–85.
Article19. Billah S, Stewart J, Staerkel G, Chen S, Gong Y, Guo M. EGFR and KRAS mutations in lung carcinoma: molecular testing by using cytology specimens. Cancer Cytopathol. 2011; 119:111–7.20. Malapelle U, Bellevicine C, De Luca C, et al. EGFR mutations detected on cytology samples by a centralized laboratory reliably predict response to gefitinib in non-small cell lung carcinoma patients. Cancer Cytopathol. 2013; 121:552–60.21. Rekhtman N, Brandt SM, Sigel CS, et al. Suitability of thoracic cytology for new therapeutic paradigms in non-small cell lung carcinoma: high accuracy of tumor subtyping and feasibility of EGFR and KRAS molecular testing. J Thorac Oncol. 2011; 6:451–8.
Article22. Sun PL, Jin Y, Kim H, Lee CT, Jheon S, Chung JH. High concordance of EGFR mutation status between histologic and corresponding cytologic specimens of lung adenocarcinomas. Cancer Cytopathol. 2013; 121:311–9.23. Lindeman NI, Cagle PT, Beasley MB, et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. Arch Pathol Lab Med. 2013; 137:828–60.
Article24. Minca EC, Lanigan CP, Reynolds JP, et al. ALK status testing in nonsmall-cell lung carcinoma by FISH on ThinPrep slides with cytology material. J Thorac Oncol. 2014; 9:464–8.
Article25. Betz BL, Dixon CA, Weigelin HC, Knoepp SM, Roh MH. The use of stained cytologic direct smears for ALK gene rearrangement analysis of lung adenocarcinoma. Cancer Cytopathol. 2013; 121:489–99.26. Proietti A, Ali G, Pelliccioni S, et al. Anaplastic lymphoma kinase gene rearrangements in cytological samples of non-small cell lung cancer: comparison with histological assessment. Cancer Cytopathol. 2014; 122:445–53.
Article27. Roy-Chowdhuri S, Aisner DL, Allen TC, et al. Biomarker testing in lung carcinoma cytology specimens: a perspective from members of the Pulmonary Pathology Society. Arch Pathol Lab Med. 2016; 140:1267–72.
Article28. Savic S, Bode B, Diebold J, et al. Detection of ALK-positive nonsmall-cell lung cancers on cytological specimens: high accuracy of immunocytochemistry with the 5A4 clone. J Thorac Oncol. 2013; 8:1004–11.
Article29. Rosenblum F, Hutchinson LM, Garver J, Woda B, Cosar E, Kurian EM. Cytology specimens offer an effective alternative to formalinfixed tissue as demonstrated by novel automated detection for ALK break-apart FISH testing and immunohistochemistry in lung adenocarcinoma. Cancer Cytopathol. 2014; 122:810–21.
Article30. Bozzetti C, Nizzoli R, Tiseo M, et al. ALK and ROS1 rearrangements tested by fluorescence in situ hybridization in cytological smears from advanced non-small cell lung cancer patients. Diagn Cytopathol. 2015; 43:941–6.31. Fernandez-Bussy S, Labarca G, Pires Y, Caviedes I, Burotto M. Molecular testing of EGFR, EGFR resistance mutation, ALK and ROS1 achieved by EBUS-TBNA in Chile. Arch Bronconeumol. 2017; 53:172–4.32. Vlajnic T, Savic S, Barascud A, et al. Detection of ROS1-positive non-small cell lung cancer on cytological specimens using immunocytochemistry. Cancer Cytopathol. 2018; 126:421–9.
Article33. Conde E, Hernandez S, Martinez R, et al. Assessment of a new ROS1 immunohistochemistry clone (SP384) for the identification of ROS1 rearrangements in patients with non-small cell lung carcinoma: the ROSING study. J Thorac Oncol. 2019; 14:2120–32.34. Sauter JL, Grogg KL, Vrana JA, Law ME, Halvorson JL, Henry MR. Young investigator challenge: validation and optimization of immunohistochemistry protocols for use on cellient cell block specimens. Cancer Cytopathol. 2016; 124:89–100.
Article35. Gruchy JR, Barnes PJ, Dakin Hache KA. CytoLyt(R) fixation and decalcification pretreatments alter antigenicity in normal tissues compared with standard formalin fixation. Appl Immunohistochem Mol Morphol. 2015; 23:297–302.
Article36. Shidham VB, Chang CC, Rao RN, Komorowski R, Chivukula M. Immunostaining of cytology smears: a comparative study to identify the most suitable method of smear preparation and fixation with reference to commonly used immunomarkers. Diagn Cytopathol. 2003; 29:217–21.
Article37. Roy-Chowdhuri S. Immunocytochemistry of cytology specimens for predictive biomarkers in lung cancer. Transl Lung Cancer Res. 2020; 9:898–905.
Article38. Chang S, Shim HS, Kim TJ, et al. Molecular biomarker testing for non-small cell lung cancer: consensus statement of the Korean Cardiopulmonary Pathology Study Group. J Pathol Transl Med. 2021; 55:181–91.
Article39. Skov BG, Skov T. Paired comparison of PD-L1 expression on cytologic and histologic specimens from malignancies in the lung assessed with PD-L1 IHC 28-8pharmDx and PD-L1 IHC 22C3pharmDx. Appl Immunohistochem Mol Morphol. 2017; 25:453–9.
Article40. Noll B, Wang WL, Gong Y, et al. Programmed death ligand 1 testing in non-small cell lung carcinoma cytology cell block and aspirate smear preparations. Cancer Cytopathol. 2018; 126:342–52.
Article41. Lozano MD, Abengozar-Muela M, Echeveste JI, et al. Programmed death-ligand 1 expression on direct Pap-stained cytology smears from non-small cell lung cancer: comparison with cell blocks and surgical resection specimens. Cancer Cytopathol. 2019; 127:470–80.
Article42. Karnes HE, Duncavage EJ, Bernadt CT. Targeted next-generation sequencing using fine-needle aspirates from adenocarcinomas of the lung. Cancer Cytopathol. 2014; 122:104–13.
Article43. Baum JE, Zhang P, Hoda RS, et al. Accuracy of next-generation sequencing for the identification of clinically relevant variants in cytology smears in lung adenocarcinoma. Cancer Cytopathol. 2017; 125:398–406.
Article44. Buttitta F, Felicioni L, Del Grammastro M, et al. Effective assessment of egfr mutation status in bronchoalveolar lavage and pleural fluids by next-generation sequencing. Clin Cancer Res. 2013; 19:691–8.45. Reynolds JP, Zhou Y, Jakubowski MA, et al. Next-generation sequencing of liquid-based cytology non-small cell lung cancer samples. Cancer Cytopathol. 2017; 125:178–87.
Article46. Treece AL, Montgomery ND, Patel NM, et al. FNA smears as a potential source of DNA for targeted next-generation sequencing of lung adenocarcinomas. Cancer Cytopathol. 2016; 124:406–14.
Article47. Roy-Chowdhuri S, Chen H, Singh RR, et al. Concurrent fine needle aspirations and core needle biopsies: a comparative study of substrates for next-generation sequencing in solid organ malignancies. Mod Pathol. 2017; 30:499–508.
Article48. Roy-Chowdhuri S, Goswami RS, Chen H, et al. Factors affecting the success of next-generation sequencing in cytology specimens. Cancer Cytopathol. 2015; 123:659–68.
Article49. Bellevicine C, Malapelle U, Vigliar E, Pisapia P, Vita G, Troncone G. How to prepare cytological samples for molecular testing. J Clin Pathol. 2017; 70:819–26.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- PD-L1 Testing in Non-small Cell Lung Cancer: Past, Present, and Future
- Personalized Medicine in Breast Cancer: A Systematic Review
- Detection of EGFR and KRAS Mutation by Pyrosequencing Analysis in Cytologic Samples of Non-Small Cell Lung Cancer
- Comparison of ThinPrep Cytology and Conventional Cytology in Bronchial Washing for Lung Cancer
- Detection of Lung Cancer using MAGE A1-6 and SSX4 RT-PCR Expression Profiles in the Bronchial Wash Fluid